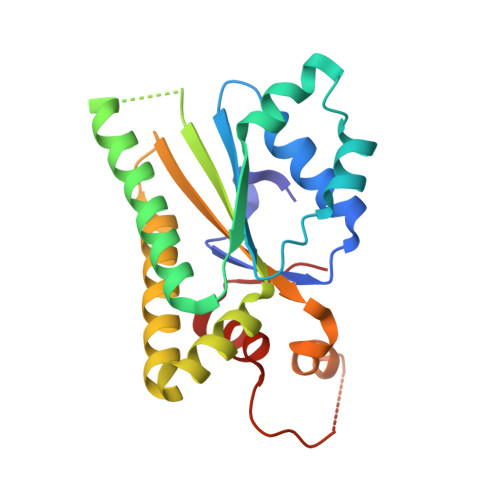
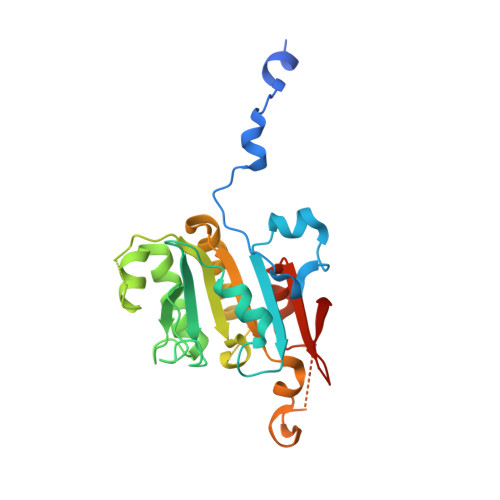
Structural and SAXS analysis of the budding yeast SHU-complex proteins
She, Z., Gao, Z.Q., Liu, Y., Wang, W.J., Liu, G.F., Shtykova, E.V., Xu, J.H., Dong, Y.H.(2012) FEBS Lett 586: 2306-2312
- PubMed: 22749910
- DOI: https://doi.org/10.1016/j.febslet.2012.06.024
- Primary Citation of Related Structures:
4EQ6 - PubMed Abstract:
In Saccharomyces cerevisiae, four proteins, Shu1, Shu2, Psy3 and Csm2, form a stable SHU-complex both in vivo and in vitro. These proteins are involved in the early stages of the homologous recombination DNA damage repair process. In this paper, the crystal structure of the Psy3-Csm2 sub-complex is presented at 1.8Å resolution and successfully fitted into our small angle X-ray scattering (SAXS) data of the SHU-complex. Taken together with our electrophoretic mobility shift assay (EMSA) results, a model is proposed for the SHU-protein complex coupled with DNA.
Organizational Affiliation:
Beijing Synchrotron Radiation Facility, Institute of High Energy Physics, Chinese Academy of Sciences, Beijing 100049, China.