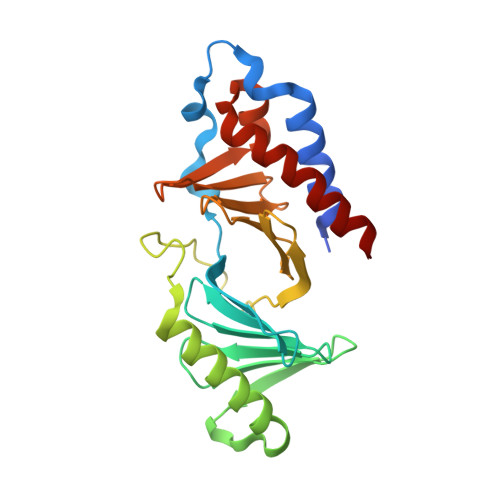
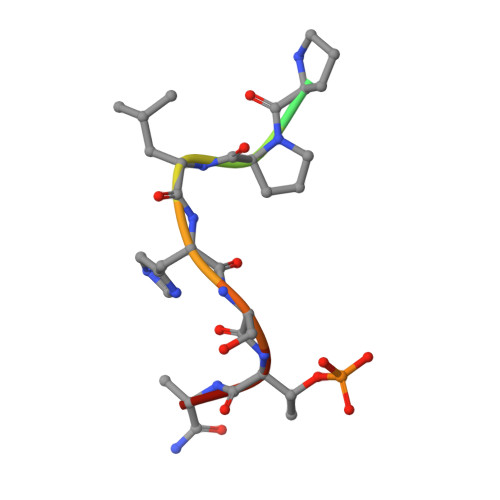
High-throughput interrogation of ligand binding mode using a fluorescence-based assay.
Sledz, P., Lang, S., Stubbs, C.J., Abell, C.(2012) Angew Chem Int Ed Engl 51: 7680-7683
- PubMed: 22730171
- DOI: https://doi.org/10.1002/anie.201202660
- Primary Citation of Related Structures:
4E9C, 4E9D - PubMed Abstract:
Probing the pocket: A high-throughput fluorescence-based thermal shift (FTS) assay utilized different forms of a protein (in gray) to establish the binding mode of a ligand (see picture). The assay serves in the rapid evaluation of structure-activity binding-mode relationships for a series of ligands of Plk1, an important target of anticancer therapy.
Organizational Affiliation:
University Chemical Laboratory, University of Cambridge, Lensfield Road, Cambridge CB2 1EW, UK.