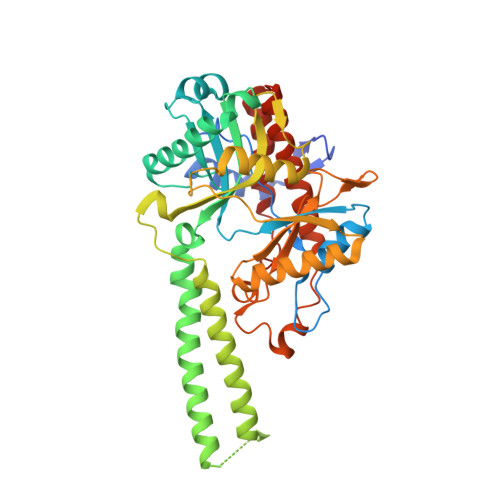
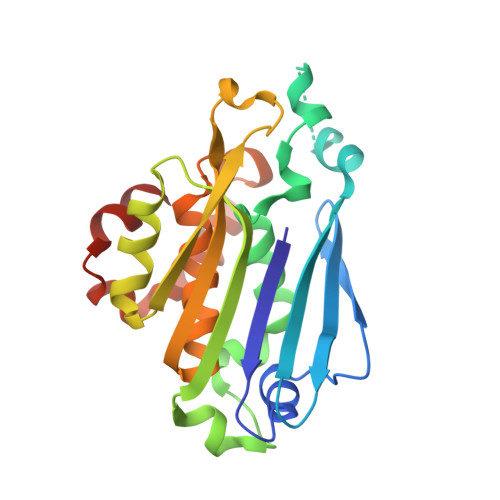
Molecular basis of bacterial protein Hen1 activating the ligase activity of bacterial protein Pnkp for RNA repair.
Wang, P., Chan, C.M., Christensen, D., Zhang, C., Selvadurai, K., Huang, R.H.(2012) Proc Natl Acad Sci U S A 109: 13248-13253
- PubMed: 22847431
- DOI: https://doi.org/10.1073/pnas.1209805109
- Primary Citation of Related Structures:
4DQZ, 4DRF, 4E6N - PubMed Abstract:
Ribotoxins cleave essential RNAs for cell killing in vivo, and the bacterial polynucleotide kinase-phosphatase (Pnkp)/hua enhancer 1 (Hen1) complex has been shown to repair ribotoxin-cleaved RNAs in vitro. Bacterial Pnkp/Hen1 is distinguished from other RNA repair systems by performing 3'-terminal 2'-O-methylation during RNA repair, which prevents the repaired RNA from repeated cleavage at the same site. To ensure the opportunity of 2'-O-methylation by bacterial Hen1 during RNA repair and, therefore, maintain the quality of the repaired RNA, Pnkp/Hen1 has evolved to require the participation of Hen1 in RNA ligation, because Pnkp alone is unable to carry out the reaction despite possessing all signature motifs of an RNA ligase. However, the precise role of Hen1 in RNA ligation is unknown. Here, we present the crystal structure of an active RNA ligase consisting of the C-terminal half of Pnkp (Pnkp-C) and the N-terminal half of Hen1 (Hen1-N) from Clostridium thermocellum. The structure reveals that the N-terminal domain of Clostridium thermocellum (Cth) Hen1, shaped like a left hand, grabs the flexible insertion module of CthPnkp and locks its conformation via further interaction with the C-terminal addition module of CthPnkp. Formation of the CthPnkp-C/Hen1-N heterodimer creates a ligation pocket with a width for two strands of RNA, depth for two nucleotides, and the adenosine monophosphate (AMP)-binding pocket at the bottom. The structure, combined with functional analyses, provides insight into the mechanism of how Hen1 activates the RNA ligase activity of Pnkp for RNA repair.
Organizational Affiliation:
Department of Biochemistry, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA.