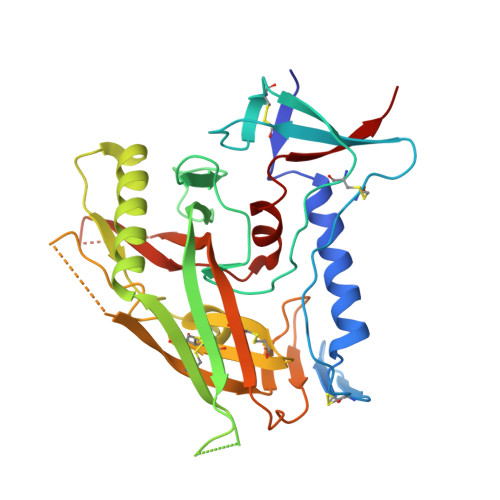
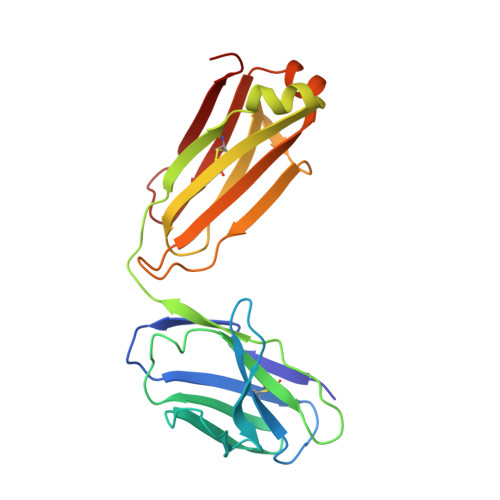
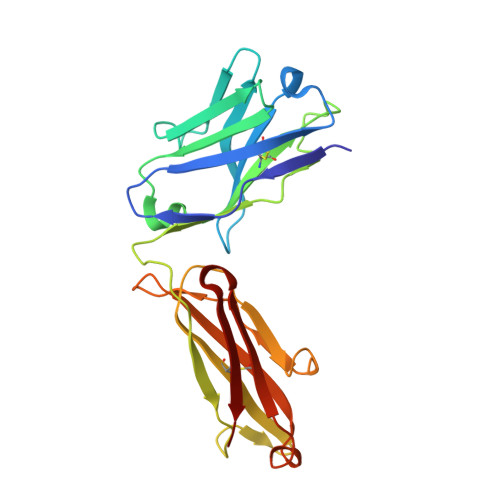
Crystal Structures of HIV-1 gp120 Envelope Glycoprotein in Complex with NBD Analogues That Target the CD4-Binding Site.
Kwon, Y.D., Lalonde, J.M., Yang, Y., Elban, M.A., Sugawara, A., Courter, J.R., Jones, D.M., Smith, A.B., Debnath, A.K., Kwong, P.D.(2014) PLoS One 9: e85940-e85940
- PubMed: 24489681
- DOI: https://doi.org/10.1371/journal.pone.0085940
- Primary Citation of Related Structures:
4DVR, 4DVS, 4DVT, 4DVV, 4DVW, 4DVX - PubMed Abstract:
Efforts to develop therapeutic agents that inhibit HIV-1 entry have led to the identification of several small molecule leads. One of the most promising is the NBD series, which binds within a conserved gp120 cavity and possesses para-halogen substituted aromatic rings, a central oxalamide linker, and a tetramethylpiperidine moiety. In this study, we characterized structurally the interactions of four NBD analogues containing meta-fluoro substitution on the aromatic ring and various heterocyclic ring replacements of the tetramethylpiperidine group. The addition of a meta-fluorine to the aromatic ring improved surface complementarity and did not alter the position of the analogue relative to gp120. By contrast, heterocyclic ring replacements of the tetramethylpiperidine moiety exhibited diverse positioning and interactions with the vestibule of the gp120 cavity. Overall, the biological profile of NBD-congeners was modulated by ligand interactions with the gp120-cavity vestibule. Herein, six co-crystal structures of NBD-analogues with gp120 provide a structural framework for continued small molecule-entry inhibitor optimization.
Organizational Affiliation:
Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, United States of America.