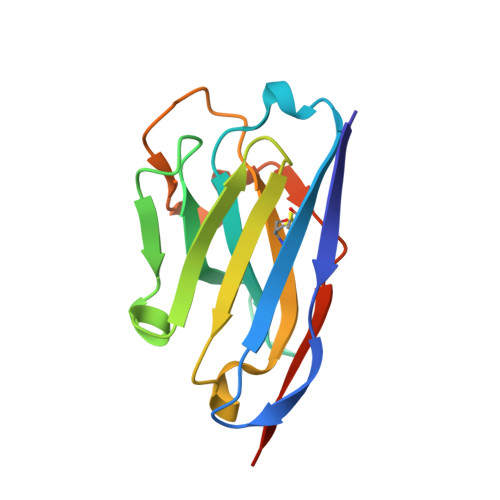
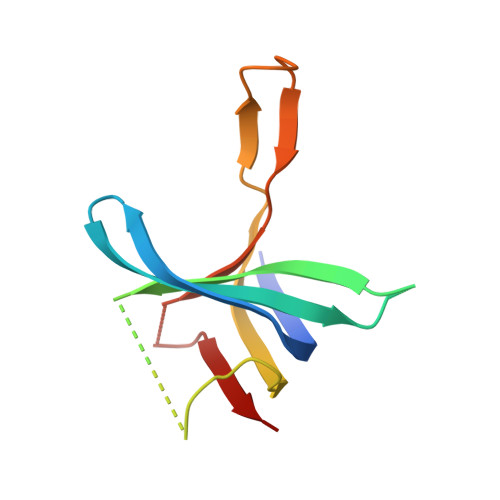
The structure of the C-terminal domain of the largest editosome interaction protein and its role in promoting RNA binding by RNA-editing ligase L2.
Park, Y.J., Budiarto, T., Wu, M., Pardon, E., Steyaert, J., Hol, W.G.(2012) Nucleic Acids Res 40: 6966-6977
- PubMed: 22561373
- DOI: https://doi.org/10.1093/nar/gks369
- Primary Citation of Related Structures:
4DK3, 4DK6, 4DKA - PubMed Abstract:
Trypanosomatids, such as the sleeping sickness parasite Trypanosoma brucei, contain a ∼ 20S RNA-editing complex, also called the editosome, which is required for U-insertion/deletion editing of mitochondrial mRNAs. The editosome contains a core of 12 proteins including the large interaction protein A1, the small interaction protein A6, and the editing RNA ligase L2. Using biochemical and structural data, we identified distinct domains of T. brucei A1 which specifically recognize A6 and L2. We provide evidence that an N-terminal domain of A1 interacts with the C-terminal domain of L2. The C-terminal domain of A1 appears to be required for the interaction with A6 and also plays a key role in RNA binding by the RNA-editing ligase L2 in trans. Three crystal structures of the C-terminal domain of A1 have been elucidated, each in complex with a nanobody as a crystallization chaperone. These structures permitted the identification of putative dsRNA recognition sites. Mutational analysis of conserved residues of the C-terminal domain identified Arg703, Arg731 and Arg734 as key requirements for RNA binding. The data show that the editing RNA ligase activity is modulated by a novel mechanism, i.e. by the trans-acting RNA binding C-terminal domain of A1.
Organizational Affiliation:
Biomolecular Structure Center, Department of Biochemistry, School of Medicine, University of Washington, Seattle, WA 98195, USA.