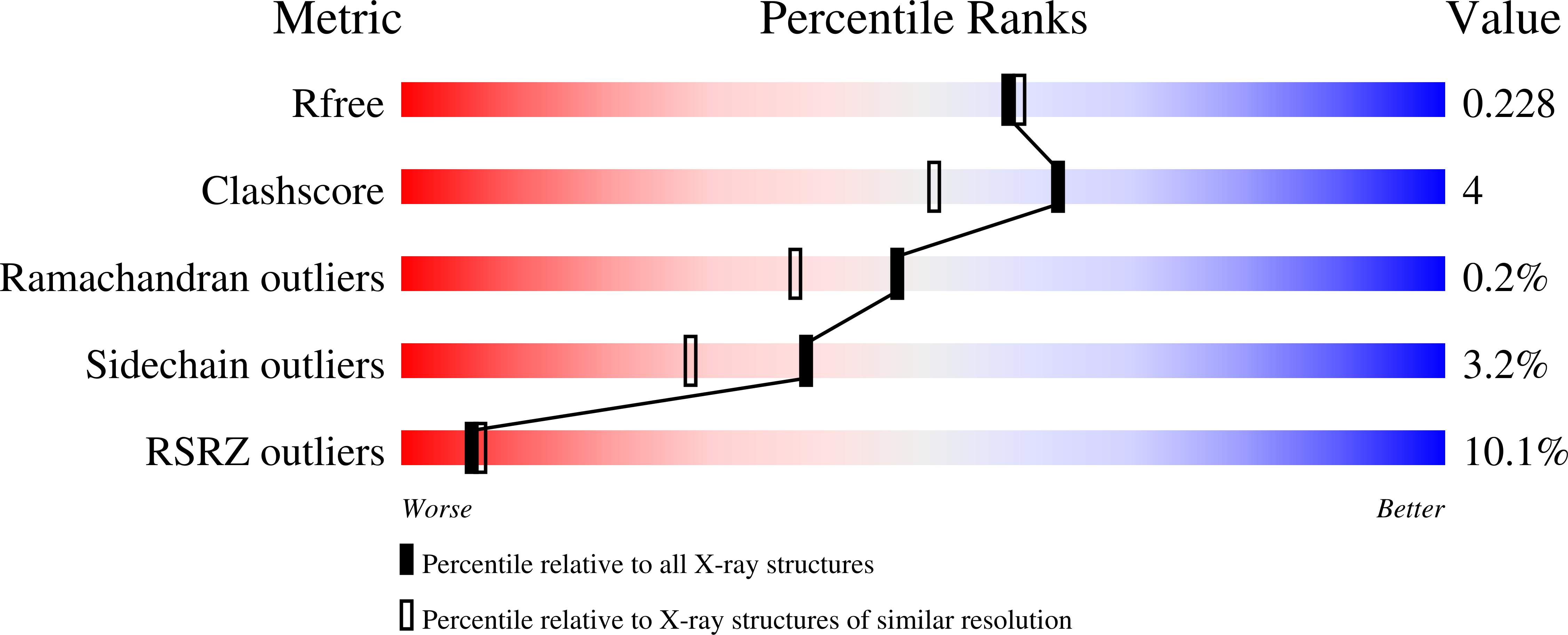
An Accessible Chiral Linker to Enhance Potency and Selectivity of Neuronal Nitric Oxide Synthase Inhibitors.
Jing, Q., Li, H., Roman, L.J., Martasek, P., Poulos, T.L., Silverman, R.B.(2014) ACS Med Chem Lett 5: 56
- PubMed: 24660051
- DOI: https://doi.org/10.1021/ml400381s
- Primary Citation of Related Structures:
4C39, 4C3A - PubMed Abstract:
The three important mammalian isozymes of nitric oxide synthase (NOS) are neuronal NOS (nNOS), endothelial NOS (eNOS), and inducible NOS (iNOS). Inhibitors of nNOS show promise as treatments for neurodegenerative diseases. Eight easily-synthesized compounds containing either one ( 20a,b ) or two ( 9a-d; 15a,b ) 2-amino-4-methylpyridine groups with a chiral pyrrolidine linker were designed as selective nNOS inhibitors. Inhibitor 9c is the best of these compounds, having a potency of 9.7 nM and dual selectivity of 693 and 295 against eNOS and iNOS, respectively. Crystal structures of nNOS complexed with either 9a or 9c show a double-headed binding mode, where each 2-aminopyridine head group interacts with either a nNOS active site Glu residue or a heme propionate. In addition, the pyrrolidine nitrogen of 9c contributes additional hydrogen bonds to the heme propionate, resulting in a unique binding orientation. In contrast, the lack of hydrogen bonds from the pyrrolidine of 9a to the heme propionate allows the inhibitor to adopt two different binding orientations. Both 9a and 9c bind to eNOS in a single-headed mode, which is the structural basis for the isozyme selectivity.
Organizational Affiliation:
Department of Chemistry, Department of Molecular Biosciences, Chemistry of Life Processes Institute, Center for Molecular Innovation and Drug Discovery, Northwestern University, Evanston, Illinois 60208-3113, USA.