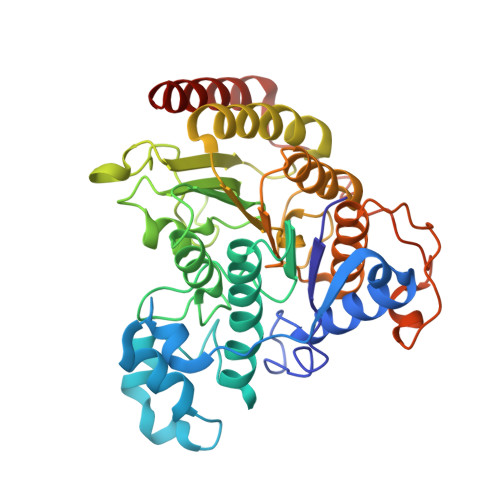
Structure of Hdac3 Bound to Co-Repressor and Inositol Tetraphosphate.
Watson, P.J., Fairall, L., Santos, G.M., Schwabe, J.W.R.(2012) Nature 481: 335
- PubMed: 22230954
- DOI: https://doi.org/10.1038/nature10728
- Primary Citation of Related Structures:
4A69 - PubMed Abstract:
Histone deacetylase enzymes (HDACs) are emerging cancer drug targets. They regulate gene expression by removing acetyl groups from lysine residues in histone tails, resulting in chromatin condensation. The enzymatic activity of most class I HDACs requires recruitment into multi-subunit co-repressor complexes, which are in turn recruited to chromatin by repressive transcription factors. Here we report the structure of a complex between an HDAC and a co-repressor, namely, human HDAC3 with the deacetylase activation domain (DAD) from the human SMRT co-repressor (also known as NCOR2). The structure reveals two remarkable features. First, the SMRT-DAD undergoes a large structural rearrangement on forming the complex. Second, there is an essential inositol tetraphosphate molecule--D-myo-inositol-(1,4,5,6)-tetrakisphosphate (Ins(1,4,5,6)P(4))--acting as an 'intermolecular glue' between the two proteins. Assembly of the complex is clearly dependent on the Ins(1,4,5,6)P(4), which may act as a regulator--potentially explaining why inositol phosphates and their kinases have been found to act as transcriptional regulators. This mechanism for the activation of HDAC3 appears to be conserved in class I HDACs from yeast to humans, and opens the way to novel therapeutic opportunities.
Organizational Affiliation:
Henry Wellcome Laboratories of Structural Biology, Department of Biochemistry, University of Leicester, Leicester LE1 9HN, UK.