Kineococcus Radiotolerans Dps Forms a Heteronuclear Mn-Fe Ferroxidase Center that May Explain the Mn-Dependent Protection Against Oxidative Stress.
Ardini, M., Fiorillo, A., Fittipaldi, M., Stefanini, S., Gatteschi, D., Llari, A., Chiancone, E.(2013) Biochim Biophys Acta 1830: 3745
- PubMed: 23396000
- DOI: https://doi.org/10.1016/j.bbagen.2013.02.003
- Primary Citation of Related Structures:
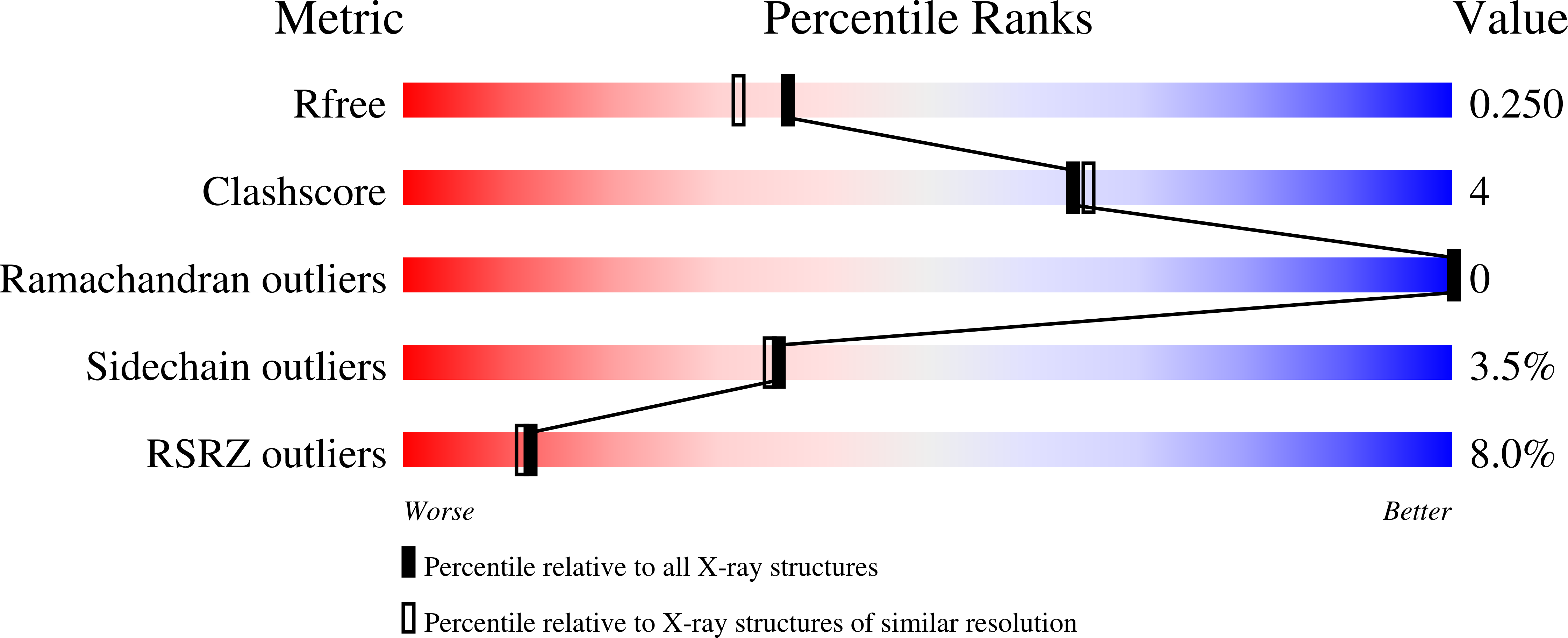
4A25 - PubMed Abstract:
The ferroxidase center of DNA-binding protein from starved cells (Dps) is a major player in the iron oxidation/detoxification process that leads to a decreased reactive oxygen species production. The possible Mn(II) participation in this process has been studied in Dps from Kineococcus radiotolerans, a radiation-resistant bacterium with a high cytosolic Mn/Fe ratio and a high capacity to survive ionizing and stress conditions. The X-ray structure of recombinant K. radiotolerans Dps loaded with Mn(II) has been solved at 2.0Å resolution. Mn(II) binding to K. radiotolerans Dps and its effect on Fe(II) oxidation have been characterized in spectroscopic measurements. In K. radiotolerans Dps, the Fe-Fe ferroxidase center can have a Mn-Fe composition. Mn(II) binds only at the high affinity, so-called A site, whereas Fe(II) binds also at the low affinity, so-called B site. The Mn-Fe and Fe-Fe centers behave distinctly upon iron oxidation by O2. A site-bound Mn(II) or Fe(II) plays a catalytic role, while B site-bound Fe(II) behaves like a substrate and can be replaced by another Fe(II) after oxidation. When H2O2 is the Fe(II) oxidant, single electrons are transferred to aromatic residues near the ferroxidase center and give rise to intra-protein radicals thereby limiting OH release in solution. The presence of the Mn-Fe center results in significant differences in the development of such intra-protein radicals. Mn(II) bound at the Dps ferroxidase center A site undergoes redox cycling provided the B site contains Fe. The results provide a likely molecular mechanism for the protective role of Mn(II) under oxidative stress conditions as it participates in redox cycling in the hetero-binuclear ferroxidase center.
Organizational Affiliation:
Department of Biochemical Sciences A. Rossi Fanelli, Sapienza University of Rome, Italy.