Conformational Stability and Crystal Packing: Polymorphism in Neurospora Crassa Cat-3
Zarate-Romero, A., Stojanoff, V., Rojas-Trejo, S.P., Hansberg, W., Rudino-Pinera, E.(2013) Acta Crystallogr Sect F Struct Biol Cryst Commun 69: 753
- PubMed: 23832201
- DOI: https://doi.org/10.1107/S1744309113013468
- Primary Citation of Related Structures:
3ZJ4, 3ZJ5, 4BIM - PubMed Abstract:
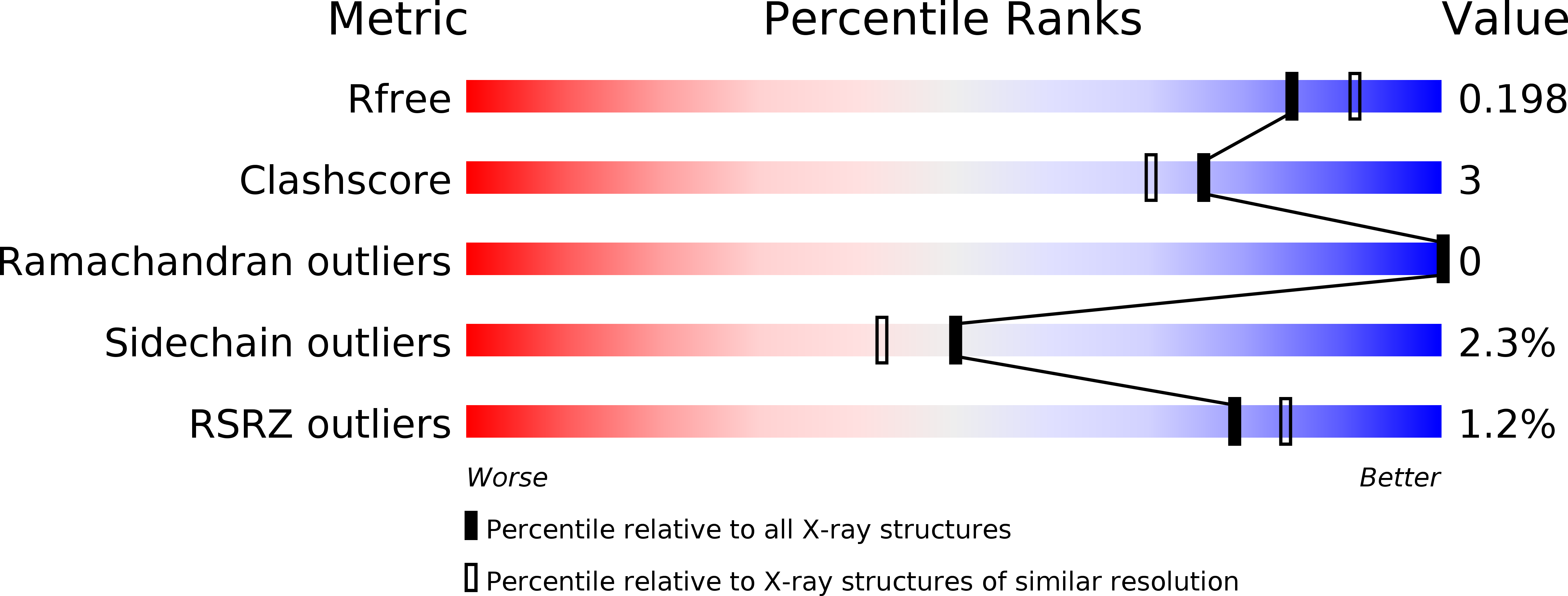
Polymorphism is frequently observed from different crystallization conditions. In proteins, the effect on conformational variability is poorly documented, with only a few reported examples. Here, three polymorphic crystal structures determined for a large-subunit catalase, CAT-3 from Neurospora crassa, are reported. Two of them belonged to new space groups, P1 and P43212, and a third structure belonged to the same space group, P212121, as the previously deposited 2.3 Å resolution structure (PDB entry 3ej6), but had a higher resolution (1.95 Å). Comparisons between these polymorphic structures highlight the conformational stability of tetrameric CAT-3 and reveal a distortion in the tetrameric structure that has not previously been described.
Organizational Affiliation:
Departamento de Medicina Molecular y Bioprocesos, Instituto de Biotecnología, Universidad Nacional Autónoma de México (UNAM), Avenida Universidad 2001, Chamilpa, 62210 Cuernavaca, MOR, Mexico. anzaro@ibt.unam.mx