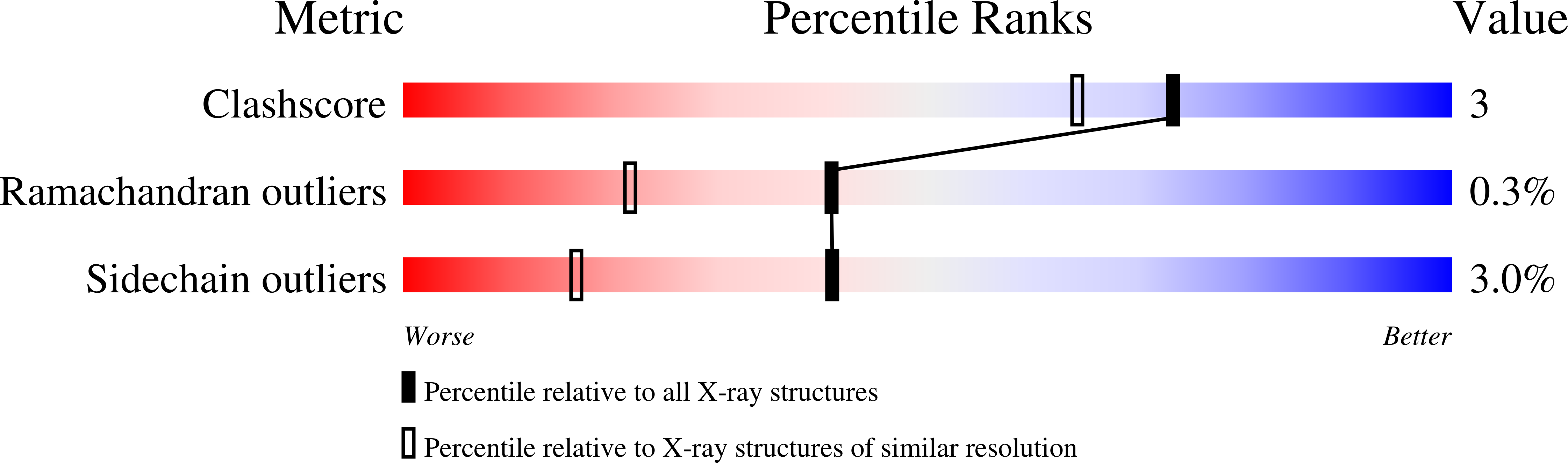A metal-mediated hydride shift mechanism for xylose isomerase based on the 1.6 A Streptomyces rubiginosus structures with xylitol and D-xylose.
Whitlow, M., Howard, A.J., Finzel, B.C., Poulos, T.L., Winborne, E., Gilliland, G.L.(1991) Proteins 9: 153-173
- PubMed: 2006134
- DOI: https://doi.org/10.1002/prot.340090302
- Primary Citation of Related Structures:
1XIS, 2XIS, 3XIS, 4XIS - PubMed Abstract:
The crystal structure of recombinant Streptomyces rubiginosus D-xylose isomerase (D-xylose keto-isomerase, EC 5.3.1.5) solved by the multiple isomorphous replacement technique has been refined to R = 0.16 at 1.64 A resolution. As observed in an earlier study at 4.0 A (Carrell et al., J. Biol. Chem. 259: 3230-3236, 1984), xylose isomerase is a tetramer composed of four identical subunits. The monomer consists of an eight-stranded parallel beta-barrel surrounded by eight helices with an extended C-terminal tail that provides extensive contacts with a neighboring monomer. The active site pocket is defined by an opening in the barrel whose entrance is lined with hydrophobic residues while the bottom of the pocket consists mainly of glutamate, aspartate, and histidine residues coordinated to two manganese ions. The structures of the enzyme in the presence of MnCl2, the inhibitor xylitol, and the substrate D-xylose in the presence and absence of MnCl2 have also been refined to R = 0.14 at 1.60 A, R = 0.15 at 1.71 A, R = 0.15 at 1.60 A, and R = 0.14 at 1.60 A, respectively. Both the ring oxygen of the cyclic alpha-D-xylose and its C1 hydroxyl are within hydrogen bonding distance of NE2 of His-54 in the structure crystallized in the presence of D-xylose. Both the inhibitor, xylitol, and the extended form of the substrate, D-xylose, bind such that the C2 and C4 OH groups interact with one of the two divalent cations found in the active site and the C1 OH with the other cation. The remainder of the OH groups hydrogen bond with neighboring amino acid side chains. A detailed mechanism for D-xylose isomerase is proposed. Upon binding of cyclic alpha-D-xylose to xylose isomerase, His-54 acts as the catalytic base in a ring opening reaction. The ring opening step is followed by binding of D-xylose, involving two divalent cations, in an extended conformation. The isomerization of D-xylose to D-xylulose involves a metal-mediated 1,2-hydride shift. The final step in the mechanism is a ring closure to produce alpha-D-xylulose. The ring closing is the reverse of the ring opening step. This mechanism accounts for the majority of xylose isomerase's biochemical properties, including (1) the lack of solvent exchange between the 2-position of D-xylose and the 1-pro-R position of D-xylulose, (2) the chemical modification of histidine and lysine, (3) the pH vs. activity profile, and (4) the requirement for two divalent cations in the mechanism.
Organizational Affiliation:
Department of Protein Engineering, Genex Corporation, Gaithersburg, Maryland 20877.