Protein-pyridinol thioester precursor for biosynthesis of the organometallic acyl-iron ligand in [Fe]-hydrogenase cofactor
Fujishiro, T., Kahnt, J., Ermler, U., Shima, S.(2015) Nat Commun 6: 6895-6895
- PubMed: 25882909
- DOI: https://doi.org/10.1038/ncomms7895
- Primary Citation of Related Structures:
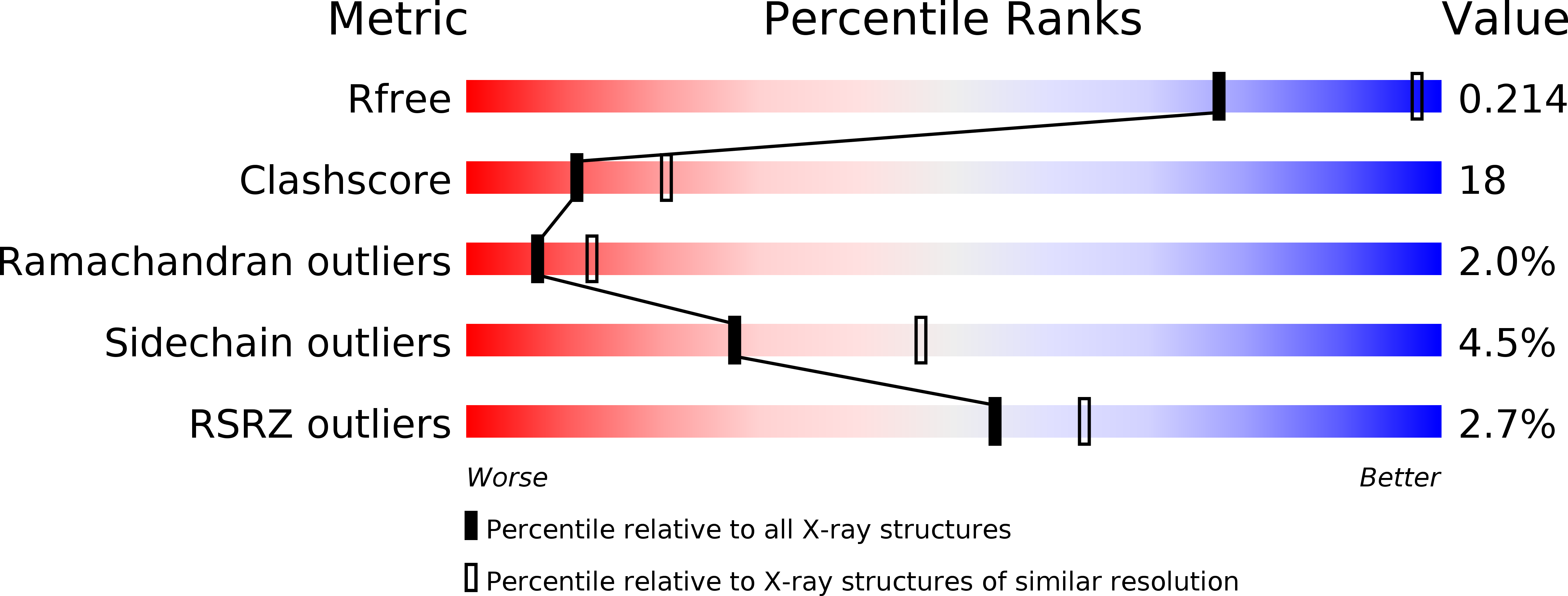
3WV7, 3WV8, 3WV9, 3WVA, 3WVB, 3WVC - PubMed Abstract:
The iron-guanylylpyridinol (FeGP) cofactor of [Fe]-hydrogenase contains a prominent iron centre with an acyl-Fe bond and is the only acyl-organometallic iron compound found in nature. Here, we identify the functions of HcgE and HcgF, involved in the biosynthesis of the FeGP cofactor using structure-to-function strategy. Analysis of the HcgE and HcgF crystal structures with and without bound substrates suggest that HcgE catalyses the adenylylation of the carboxy group of guanylylpyridinol (GP) to afford AMP-GP, and subsequently HcgF catalyses the transesterification of AMP-GP to afford a Cys (HcgF)-S-GP thioester. Both enzymatic reactions are confirmed by in vitro assays. The structural data also offer plausible catalytic mechanisms. This strategy of thioester activation corresponds to that used for ubiquitin activation, a key event in the regulation of multiple cellular processes. It further implicates a nucleophilic attack onto the acyl carbon presumably via an electron-rich Fe(0)- or Fe(I)-carbonyl complex in the Fe-acyl formation.
Organizational Affiliation:
Max Planck Institute for Terrestrial Microbiology, Karl-von-Frisch-Strasse 10, 35043 Marburg, Germany.