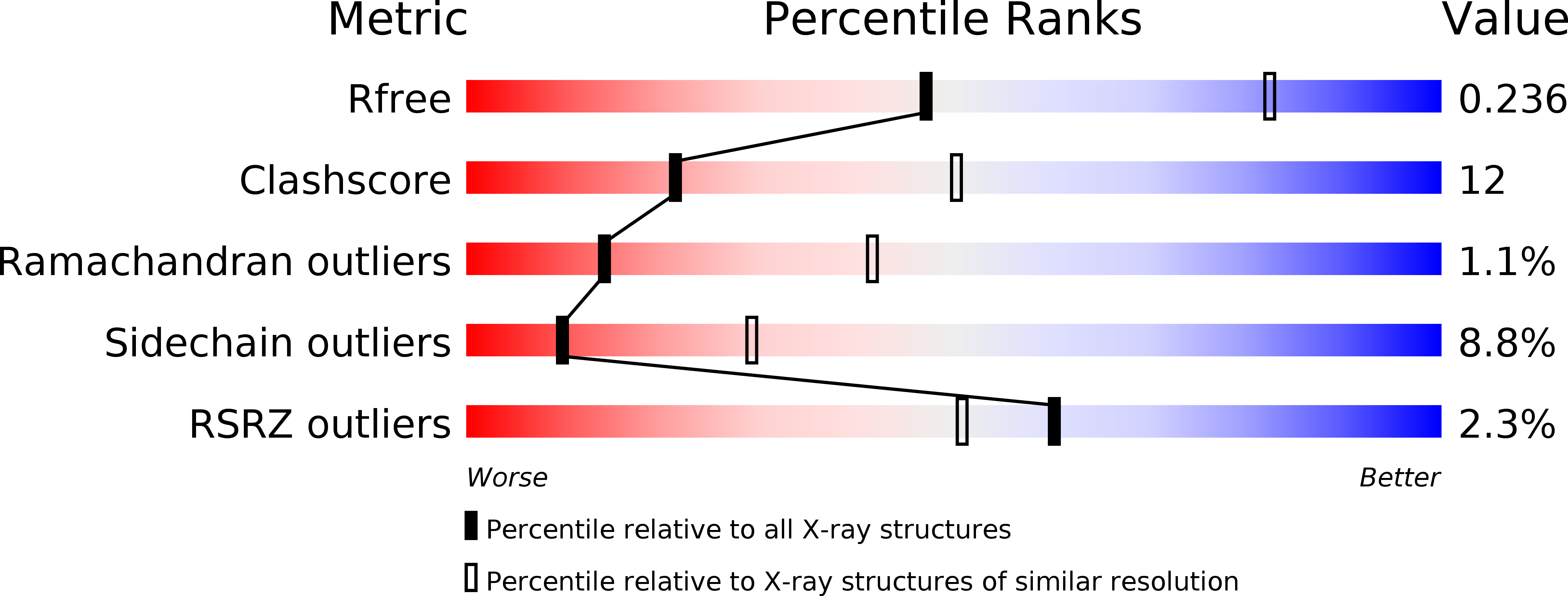
Crystal structure of Bombyx mori arylphorins reveals a 3:3 heterohexamer with multiple papain cleavage sites
Hou, Y., Li, J., Li, Y., Dong, Z., Xia, Q., Yuan, Y.A.(2014) Protein Sci 23: 735-746
- PubMed: 24639361
- DOI: https://doi.org/10.1002/pro.2457
- Primary Citation of Related Structures:
3WJM - PubMed Abstract:
In holometabolous insects, the accumulation and utilization of storage proteins (SPs), including arylphorins and methionine-rich proteins, are critical for the insect metamorphosis. SPs function as amino acids reserves, which are synthesized in fat body, secreted into the larval hemolymph and taken up by fat body shortly before pupation. However, the detailed molecular mechanisms of digestion and utilization of SPs during development are largely unknown. Here, we report the crystal structure of Bombyx mori arylphorins at 2.8 Å, which displays a heterohexameric structural arrangement formed by trimerization of dimers comprising two structural similar arylphorins. Our limited proteolysis assay and microarray data strongly suggest that papain-like proteases are the major players for B. mori arylphorins digestion in vitro and in vivo. Consistent with the biochemical data, dozens of papain cleavage sites are mapped on the surface of the heterohexameric structure of B. mori arylphorins. Hence, our results provide the insightful information to understand the metamorphosis of holometabolous insects at molecular level.
Organizational Affiliation:
State Key Laboratory of Silkworm Genome Biology, College of Biotechnology, Southwest University, Beibei, Chongqing, 400715, China; Department of Biological Sciences and Center for Bioimaging Sciences, National University of Singapore, Singapore, 117543, Singapore; SWU-NUS Joint Laboratory in Structural Genomics, Southwest University, Beibei, Chongqing, 400715, China.