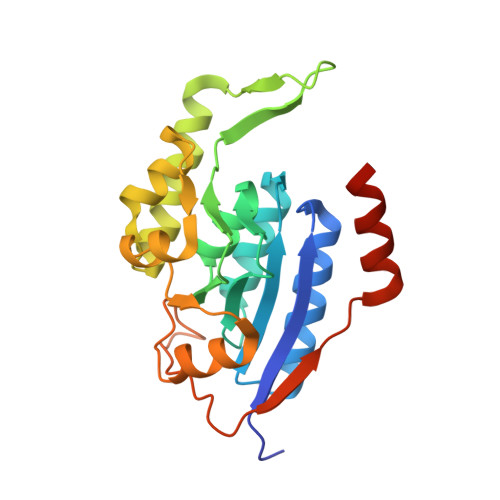
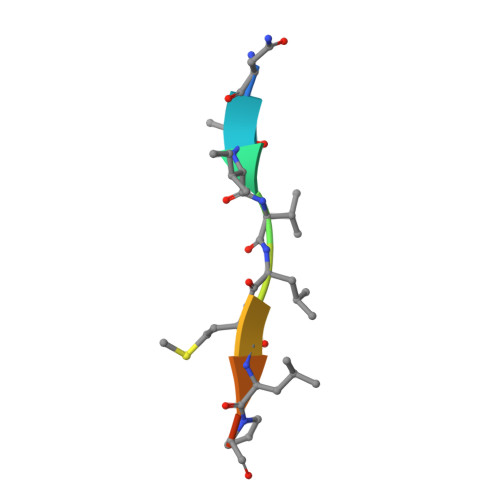
Structural and biochemical analysis of a thermostable membrane-bound stomatin-specific protease.
Yokoyama, H., Kobayashi, D., Takizawa, N., Fujii, S., Matsui, I.(2013) J Synchrotron Radiat 20: 933-937
- PubMed: 24121343
- DOI: https://doi.org/10.1107/S0909049513021328
- Primary Citation of Related Structures:
3WG5 - PubMed Abstract:
Membrane-bound proteases are involved in various regulatory functions. The N-terminal region of PH1510p (1510-N) from the hyperthermophilic archaeon Pyrococcus horikoshii is a serine protease with a catalytic Ser-Lys dyad (Ser97 and Lys138), and specifically cleaves the C-terminal hydrophobic region of the p-stomatin PH1511p. In a form of human hemolytic anemia known as hereditary stomatocytosis, the stomatin protein is deficient in the erythrocyte membrane due to mis-trafficking. In order to understand the catalytic mechanism of 1510-N in more detail, here the structural and biochemical analysis of 1510-N is reported. Two degraded products were produced via acyl-enzyme intermediates. 1510-N is a thermostable protease, and thus crystallization after heat treatment of the protease-peptide complex was attempted in order to understand the catalytic mechanism of 1510-N. The structure after heat treatment is almost identical to that with no heat treatment. According to the superposition between the structures with heat treatment and with no heat treatment, the N-terminal half of the peptide is superposed well, whereas the C-terminal half of the peptide is slightly deviated. The N-terminal half of the peptide binds to 1510-N more tightly than the C-terminal half of the peptide. The flexible L2 loops of 1510-N cover the peptide, and are involved in the protease activity.
Organizational Affiliation:
School of Pharmaceutical Sciences, University of Shizuoka, 52-1 Yada, Suruga-ku, Shizuoka 422-8526, Japan.