Impaired alpha-TTP-PIPs interaction underlies familial vitamin E deficiency
Kono, N., Ohto, U., Hiramatsu, T., Urabe, M., Uchida, Y., Satow, Y., Arai, H.(2013) Science 340: 1106-1110
- PubMed: 23599266
- DOI: https://doi.org/10.1126/science.1233508
- Primary Citation of Related Structures:
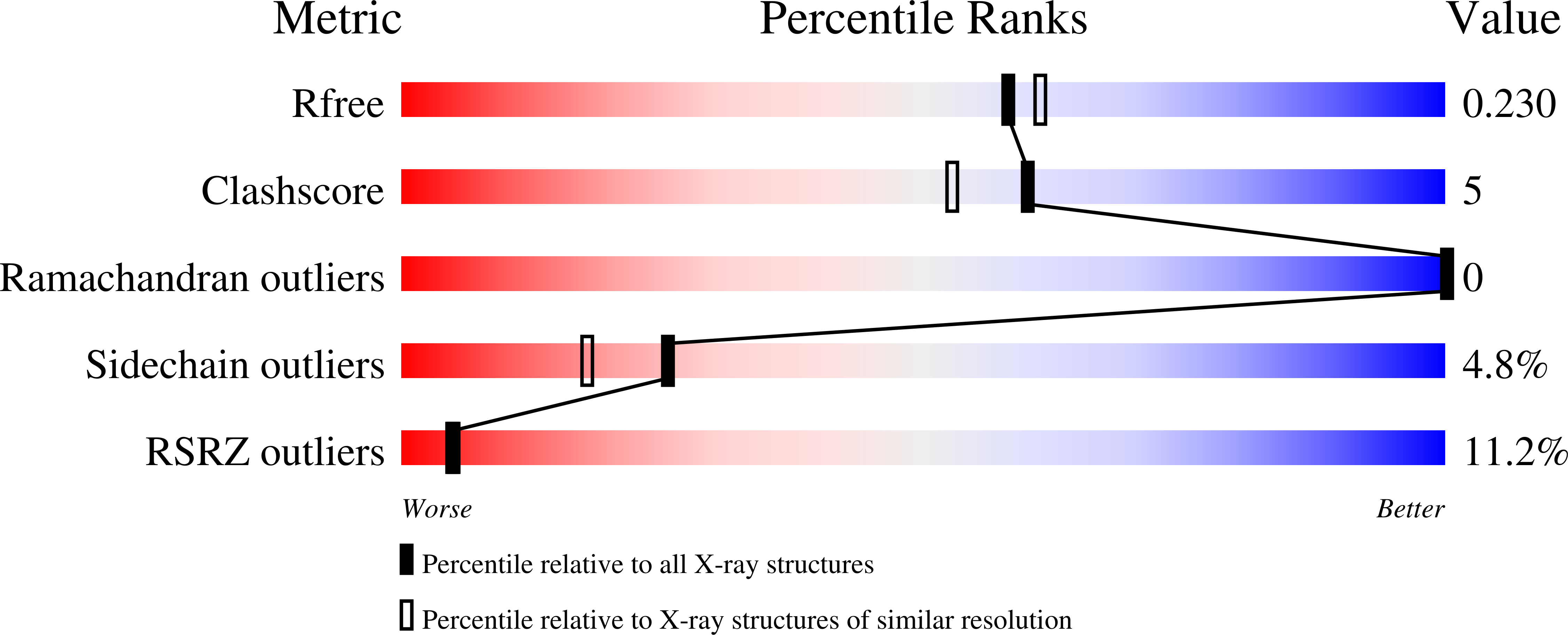
3W67, 3W68 - PubMed Abstract:
α-Tocopherol (vitamin E) transfer protein (α-TTP) regulates the secretion of α-tocopherol from liver cells. Missense mutations of some arginine residues at the surface of α-TTP cause severe vitamin E deficiency in humans, but the role of these residues is unclear. Here, we found that wild-type α-TTP bound phosphatidylinositol phosphates (PIPs), whereas the arginine mutants did not. In addition, PIPs in the target membrane promoted the intermembrane transfer of α-tocopherol by α-TTP. The crystal structure of the α-TTP-PIPs complex revealed that the disease-related arginine residues interacted with phosphate groups of the PIPs and that the PIPs binding caused the lid of the α-tocopherol-binding pocket to open. Thus, PIPs have a role in promoting the release of a ligand from a lipid-transfer protein.
Organizational Affiliation:
Graduate School of Pharmaceutical Sciences, University of Tokyo, Tokyo, Japan.