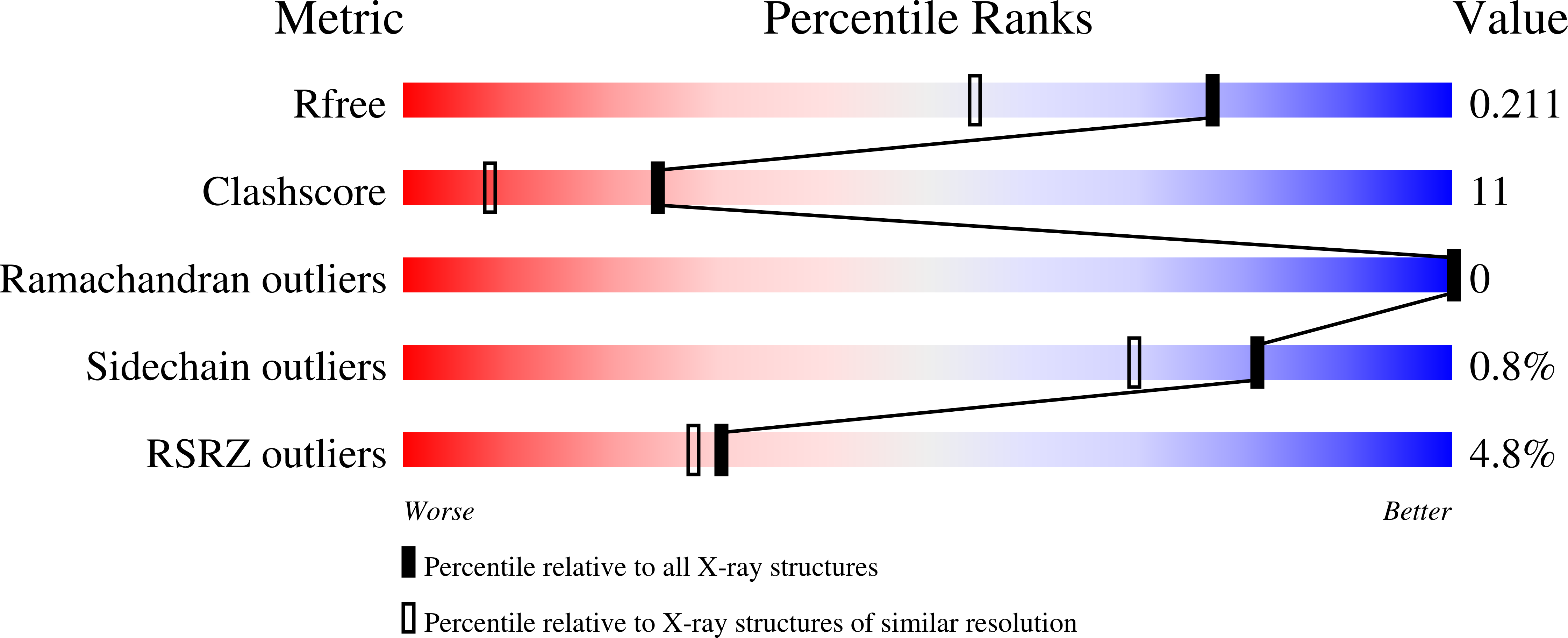
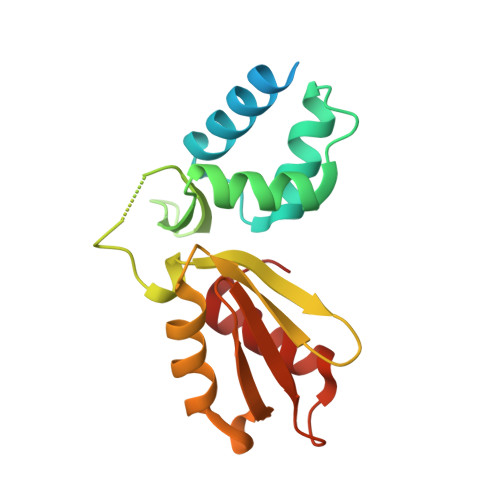
1.60 Angstrom resolution crystal structure of an arginine repressor from Vibrio vulnificus CMCP6
Halavaty, A.S., Minasov, G., Filippova, E., Shuvalova, L., Winsor, J., Dubrovska, I., Peterson, S., Anderson, W.F., Center for Structural Genomics of Infectious Diseases (CSGID)To be published.