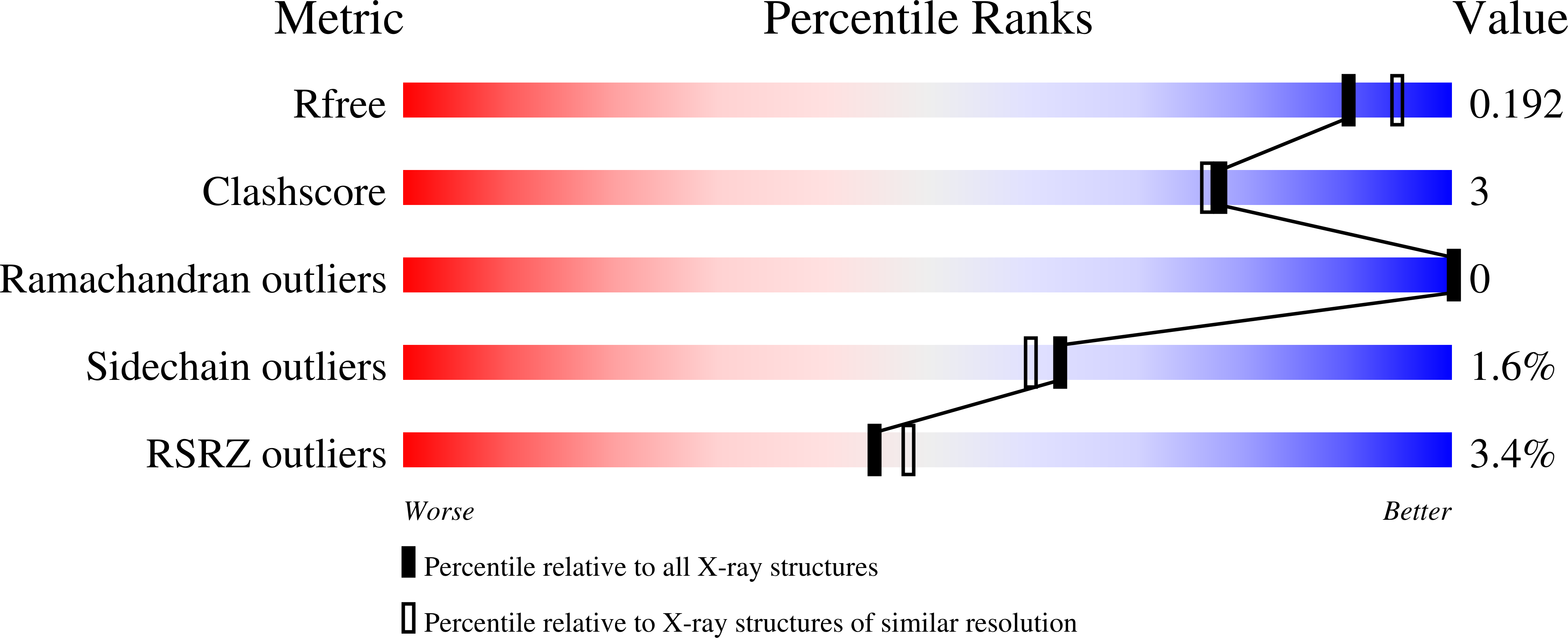
Base-modified Donor Analogues Reveal Novel Dynamic Features of a Glycosyltransferase.
Jrgensen, R., Pesnot, T., Lee, H.J., Palcic, M.M., Wagner, G.K.(2013) J Biol Chem 288: 26201-26208
- PubMed: 23836908
- DOI: https://doi.org/10.1074/jbc.M113.465963
- Primary Citation of Related Structures:
3V0L, 3V0M, 3V0N, 3V0O, 3V0P, 3V0Q - PubMed Abstract:
Glycosyltransferases (GTs) are enzymes that are involved, as Nature's "glycosylation reagents," in many fundamental biological processes including cell adhesion and blood group biosynthesis. Although of similar importance to that of other large enzyme families such as protein kinases and proteases, the undisputed potential of GTs for chemical biology and drug discovery has remained largely unrealized to date. This is due, at least in part, to a relative lack of GT inhibitors and tool compounds for structural, mechanistic, and cellular studies. In this study, we have used a novel class of GT donor analogues to obtain new structural and enzymological information for a representative blood group GT. These analogues interfere with the folding of an internal loop and the C terminus, which are essential for catalysis. Our experiments have led to the discovery of an entirely new active site folding mode for this enzyme family, which can be targeted in inhibitor development, similar to the DFG motif in protein kinases. Taken together, our results provide new insights into substrate binding, dynamics, and utilization in this important enzyme family, which can very likely be harnessed for the rational development of new GT inhibitors and probes.
Organizational Affiliation:
From the Department of Microbiology and Infection Control, Statens Serum Institut, DK-2300 Copenhagen S, Denmark,; the Carlsberg Laboratory, Gamle Carlsberg Vej 10, DK-1799 Copenhagen V, Denmark,. Electronic address: renj@ssi.dk.