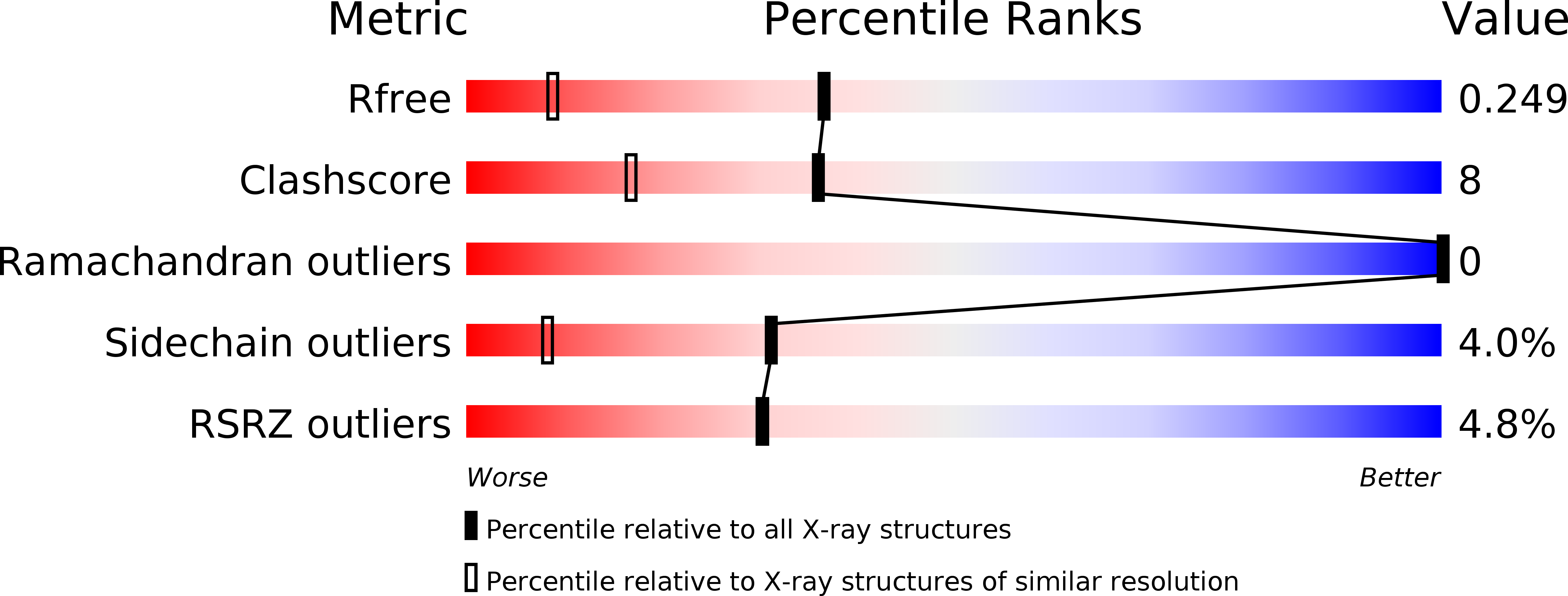
Structural basis for the antifreeze activity of an ice-binding protein from an Arctic yeast.
Lee, J.H., Park, A.K., Do, H., Park, K.S., Moh, S.H., Chi, Y.M., Kim, H.J.(2012) J Biol Chem
- PubMed: 22303017
- DOI: https://doi.org/10.1074/jbc.M111.331835
- Primary Citation of Related Structures:
3UYU, 3UYV - PubMed Abstract:
Arctic yeast Leucosporidium sp. produces a glycosylated ice-binding protein (LeIBP) with a molecular mass of ∼25 kDa, which can lower the freezing point below the melting point once it binds to ice. LeIBP is a member of a large class of ice-binding proteins, the structures of which are unknown. Here, we report the crystal structures of non-glycosylated LeIBP and glycosylated LeIBP at 1.57- and 2.43-Å resolution, respectively. Structural analysis of the LeIBPs revealed a dimeric right-handed β-helix fold, which is composed of three parts: a large coiled structural domain, a long helix region (residues 96-115 form a long α-helix that packs along one face of the β-helix), and a C-terminal hydrophobic loop region ((243)PFVPAPEVV(251)). Unexpectedly, the C-terminal hydrophobic loop region has an extended conformation pointing away from the body of the coiled structural domain and forms intertwined dimer interactions. In addition, structural analysis of glycosylated LeIBP with sugar moieties attached to Asn(185) provides a basis for interpreting previous biochemical analyses as well as the increased stability and secretion of glycosylated LeIBP. We also determined that the aligned Thr/Ser/Ala residues are critical for ice binding within the B face of LeIBP using site-directed mutagenesis. Although LeIBP has a common β-helical fold similar to that of canonical hyperactive antifreeze proteins, the ice-binding site is more complex and does not have a simple ice-binding motif. In conclusion, we could identify the ice-binding site of LeIBP and discuss differences in the ice-binding modes compared with other known antifreeze proteins and ice-binding proteins.
Organizational Affiliation:
Division of Polar Life Sciences, Korea Polar Research Institute, Incheon 406-840, Republic of Korea.