Crystal structure of Butea monosperma seed lectin
Abhilash, J., Geethanandan, K., Bharath, S.R., Sadasivan, C., Haridas, M.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Lectin Alpha chain | 256 | Butea monosperma | Mutation(s): 0 | 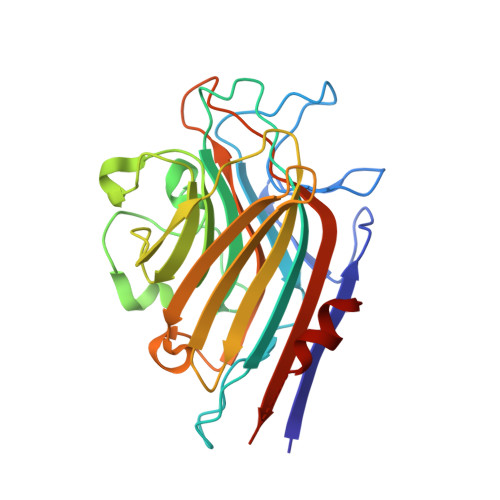 | |
UniProt | |||||
Find proteins for H2L2M6 (Butea monosperma) Explore H2L2M6 Go to UniProtKB: H2L2M6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | H2L2M6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Lectin Beta Chain | 242 | Butea monosperma | Mutation(s): 0 | 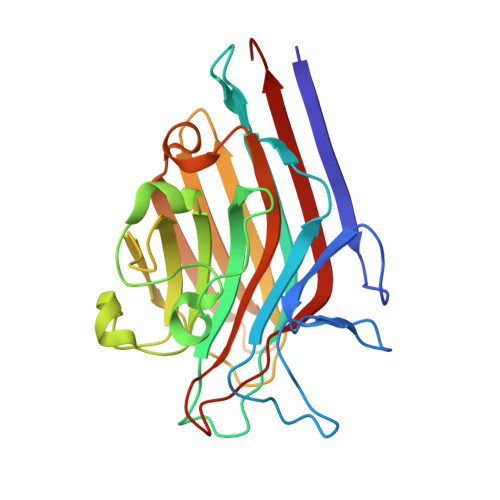 | |
UniProt | |||||
Find proteins for H2L2M6 (Butea monosperma) Explore H2L2M6 Go to UniProtKB: H2L2M6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | H2L2M6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose | I, K, L | 4 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G18638YB GlyCosmos: G18638YB GlyGen: G18638YB | |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose | J | 5 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G17689EW GlyCosmos: G17689EW GlyGen: G17689EW | |||||
| Ligands 6 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG Query on NAG | FA [auth D] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| XYP Query on XYP | W [auth B] | beta-D-xylopyranose C5 H10 O5 SRBFZHDQGSBBOR-KKQCNMDGSA-N |  | ||
| ABU Query on ABU | AA [auth C] EA [auth D] JA [auth E] LA [auth E] OA [auth F] | GAMMA-AMINO-BUTANOIC ACID C4 H9 N O2 BTCSSZJGUNDROE-UHFFFAOYSA-N |  | ||
| GOL Query on GOL | DA [auth D] IA [auth E] KA [auth E] R [auth A] RA [auth G] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| MN Query on MN | CA [auth D] HA [auth E] NA [auth F] P [auth A] QA [auth G] | MANGANESE (II) ION Mn WAEMQWOKJMHJLA-UHFFFAOYSA-N |  | ||
| CA Query on CA | BA [auth D] GA [auth E] MA [auth F] O [auth A] PA [auth G] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 78.45 | α = 74.3 |
| b = 78.91 | β = 76.65 |
| c = 101.85 | γ = 86.88 |
| Software Name | Purpose |
|---|---|
| MAR345dtb | data collection |
| MOLREP | phasing |
| REFMAC | refinement |
| MOSFLM | data reduction |
| SCALA | data scaling |