High-Resolution Crystal Structure of Spectrin SH3 Domain Fused with a Proline-Rich Peptide.
Gushchina, L.V., Gabdulkhakov, A.G., Nikonov, S.V., Filimonov, V.V.(2011) J Biomol Struct Dyn 29: 485-495
- PubMed: 22066535
- DOI: https://doi.org/10.1080/07391102.2011.10507400
- Primary Citation of Related Structures:
3THK - PubMed Abstract:
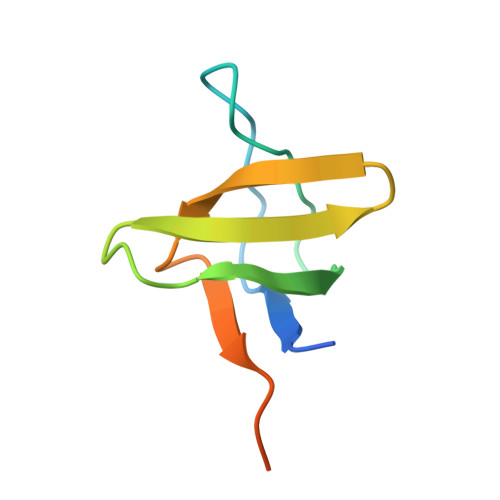
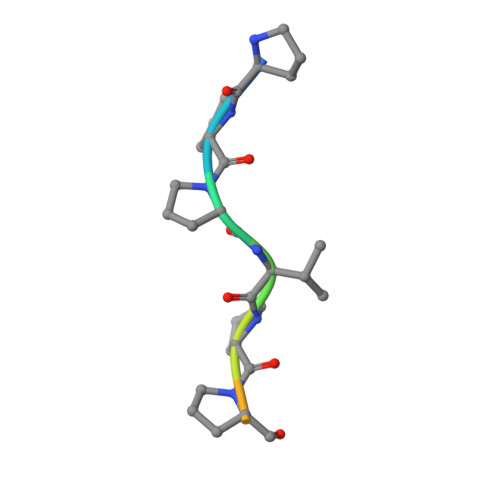
A new chimeric protein, named WT-CIIA, was designed by connecting the proline-rich decapeptide PPPVPPYSAG to the C-terminus of the alpha-spectrin SH3 domain through a natural twelve-residue linker to obtain a single-chain model that would imitate intramolecular SH3-ligand interaction. The crystal structure of this fusion protein was determined at 1.7 Å resolution. The asymmetric unit of the crystal contained two SH3 globules contacting with one PPPVPPY fragment located between them. The domains are related by the two-fold non-crystallographic axis and the ligand lies in two opposite orientations with respect to the conservative binding sites of SH3 domains.
Organizational Affiliation:
Institute of Protein Research, Russian Academy of Sciences, Pushchino, Moscow Region, Russia.