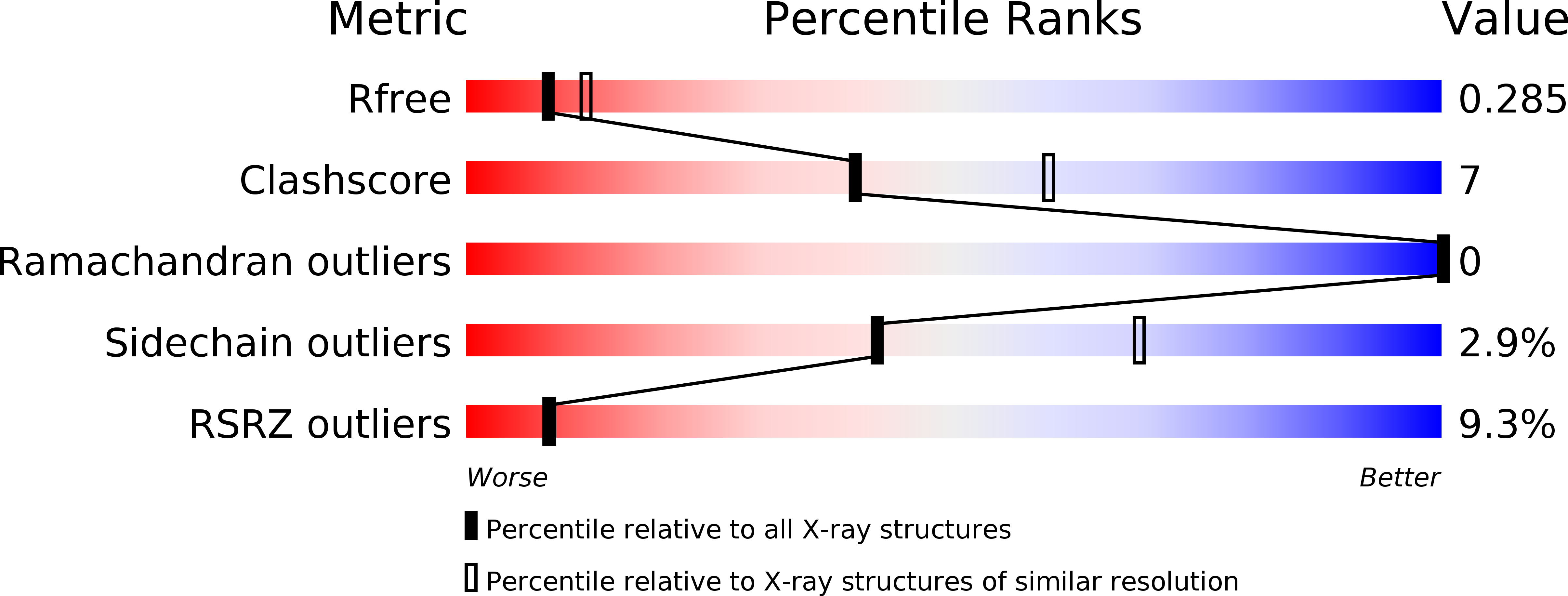
Design of novel FN3 domains with high stability by a consensus sequence approach.
Jacobs, S.A., Diem, M.D., Luo, J., Teplyakov, A., Obmolova, G., Malia, T., Gilliland, G.L., O'Neil, K.T.(2012) Protein Eng Des Sel 25: 107-117
- PubMed: 22240293
- DOI: https://doi.org/10.1093/protein/gzr064
- Primary Citation of Related Structures:
3TES, 3TEU - PubMed Abstract:
The use of consensus design to produce stable proteins has been applied to numerous structures and classes of proteins. Here, we describe the engineering of novel FN3 domains from two different proteins, namely human fibronectin and human tenascin-C, as potential alternative scaffold biotherapeutics. The resulting FN3 domains were found to be robustly expressed in Escherichia coli, soluble and highly stable, with melting temperatures of 89 and 78°C, respectively. X-ray crystallography was used to confirm that the consensus approach led to a structure consistent with the FN3 design despite having only low-sequence identity to natural FN3 domains. The ability of the Tenascin consensus domain to withstand mutations in the loop regions connecting the β-strands was investigated using alanine scanning mutagenesis demonstrating the potential for randomization in these regions. Finally, rational design was used to produce point mutations that significantly increase the stability of one of the consensus domains. Together our data suggest that consensus FN3 domains have potential utility as alternative scaffold therapeutics.
Organizational Affiliation:
Janssen Research & Development, L.L.C., Radnor, PA 19087, USA. sjacobs9@its.jnj.com