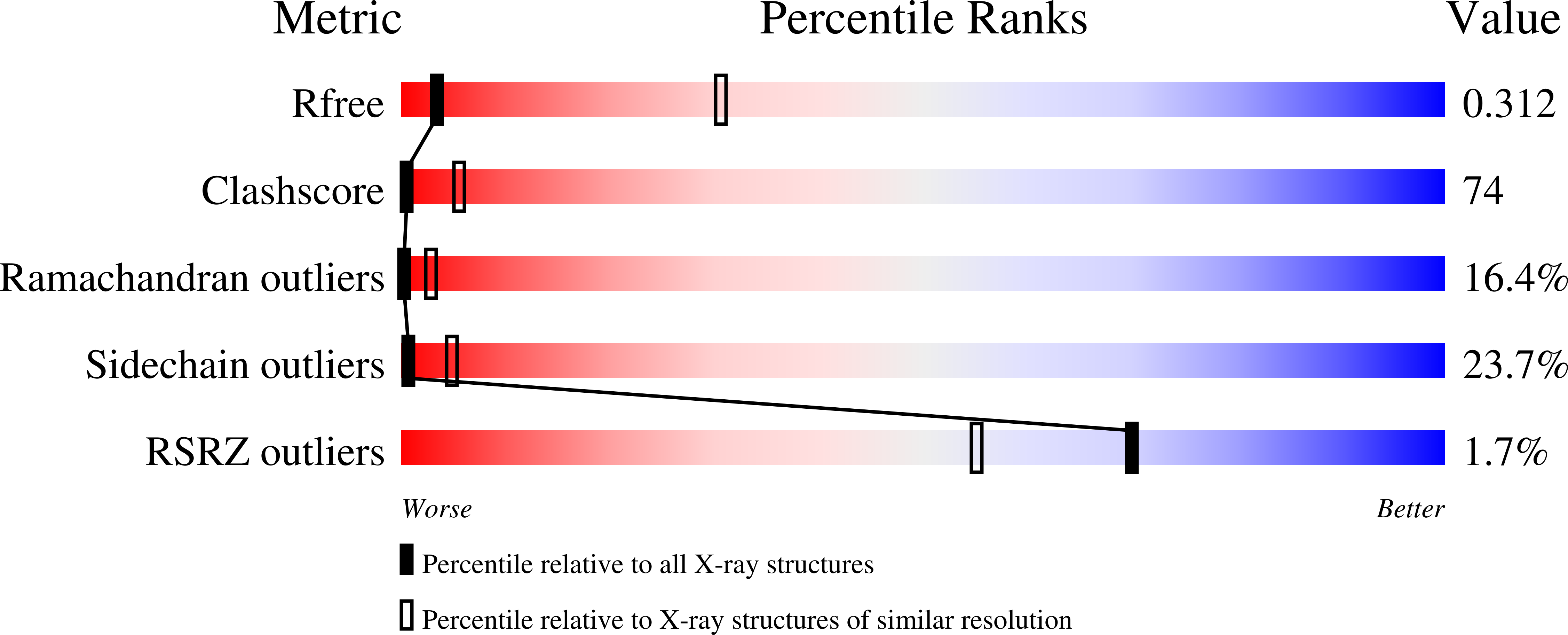
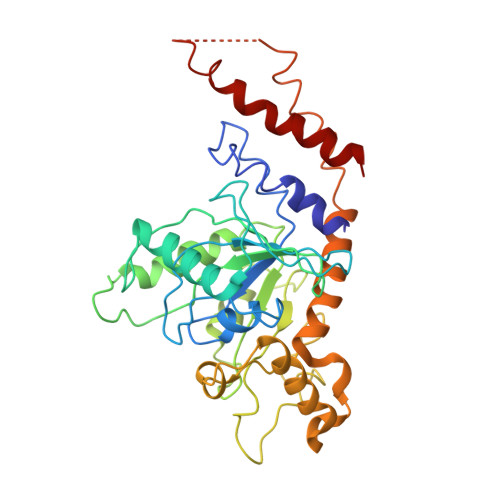
Structural basis for mechanochemical role of Arabidopsis thaliana dynamin-related protein in membrane fission
Yan, L.M., Ma, Y.Y., Sun, Y.N., Gao, J., Chen, X., Liu, J., Wang, C., Rao, Z., Lou, Z.Y.(2011) J Mol Cell Biol 3: 378-381
- PubMed: 22107825
- DOI: https://doi.org/10.1093/jmcb/mjr032
- Primary Citation of Related Structures:
3T34, 3T35