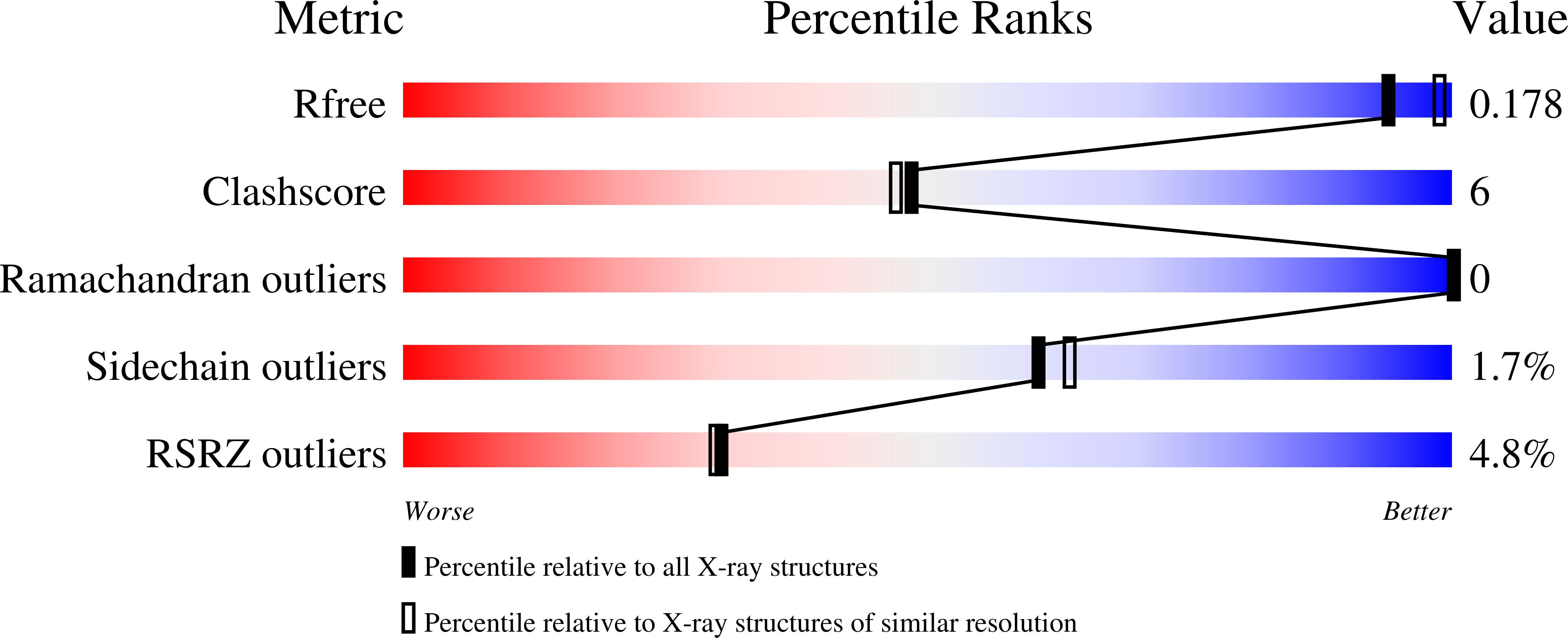
Mycobacterium tuberculosis Eis protein initiates suppression of host immune responses by acetylation of DUSP16/MKP-7
Kim, K.H., An, D.R., Song, J., Yoon, J.Y., Kim, H.S., Yoon, H.J., Im, H.N., Kim, J., Kim, D.J., Lee, S.J., Kim, K.H., Lee, H.M., Kim, H.J., Jo, E.K., Lee, J.Y., Suh, S.W.(2012) Proc Natl Acad Sci U S A 109: 7729-7734
- PubMed: 22547814
- DOI: https://doi.org/10.1073/pnas.1120251109
- Primary Citation of Related Structures:
3RYO, 3SXN, 3UY5 - PubMed Abstract:
The intracellular pathogen Mycobacterium tuberculosis (Mtb) causes tuberculosis. Enhanced intracellular survival (Eis) protein, secreted by Mtb, enhances survival of Mycobacterium smegmatis (Msm) in macrophages. Mtb Eis was shown to suppress host immune defenses by negatively modulating autophagy, inflammation, and cell death through JNK-dependent inhibition of reactive oxygen species (ROS) generation. Mtb Eis was recently demonstrated to contribute to drug resistance by acetylating multiple amines of aminoglycosides. However, the mechanism of enhanced intracellular survival by Mtb Eis remains unanswered. Therefore, we have characterized both Mtb and Msm Eis proteins biochemically and structurally. We have discovered that Mtb Eis is an efficient N(ε)-acetyltransferase, rapidly acetylating Lys55 of dual-specificity protein phosphatase 16 (DUSP16)/mitogen-activated protein kinase phosphatase-7 (MKP-7), a JNK-specific phosphatase. In contrast, Msm Eis is more efficient as an N(α)-acetyltransferase. We also show that Msm Eis acetylates aminoglycosides as readily as Mtb Eis. Furthermore, Mtb Eis, but not Msm Eis, inhibits LPS-induced JNK phosphorylation. This functional difference against DUSP16/MKP-7 can be understood by comparing the structures of two Eis proteins. The active site of Mtb Eis with a narrow channel seems more suitable for sequence-specific recognition of the protein substrate than the pocket-shaped active site of Msm Eis. We propose that Mtb Eis initiates the inhibition of JNK-dependent autophagy, phagosome maturation, and ROS generation by acetylating DUSP16/MKP-7. Our work thus provides insight into the mechanism of suppressing host immune responses and enhancing mycobacterial survival within macrophages by Mtb Eis.
Organizational Affiliation:
Department of Chemistry, College of Natural Sciences, Seoul National University, Seoul 151-742, Korea.