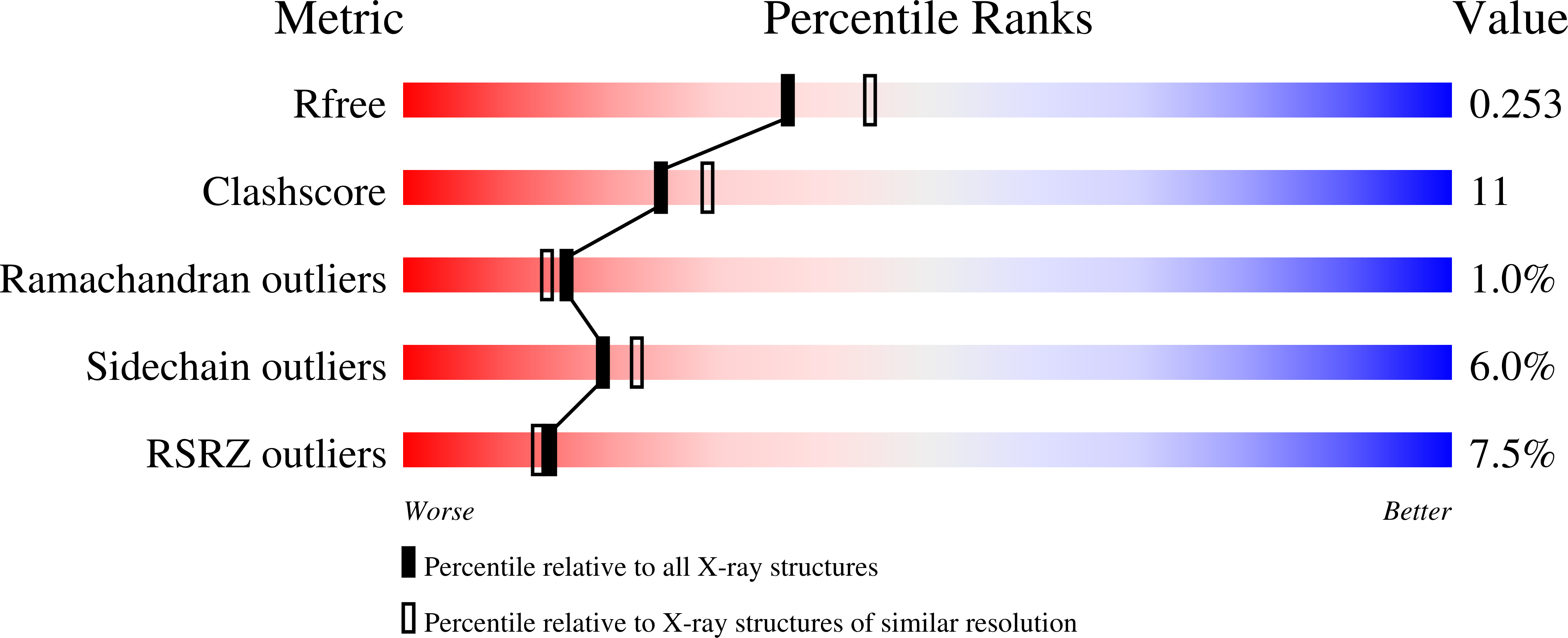
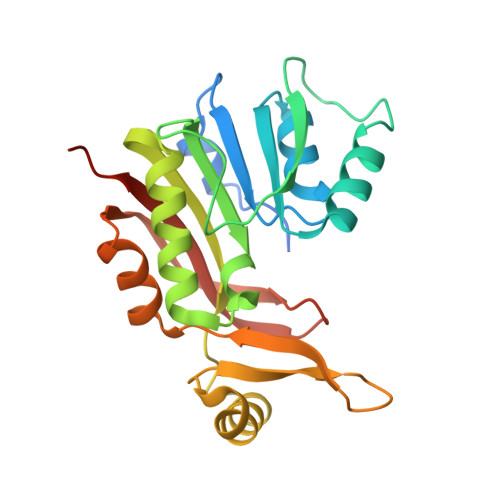
Crystal Structure of SAM-dependent methyltransferases Q8PUK2_METMA from Methanosarcina mazei.
Vorobiev, S., Neely, H., Seetharaman, J., Snyder, A., Patel, P., Xiao, R., Ciccosanti, C., Everett, J.K., Nair, R., Acton, T.B., Rost, B., Montelione, G.T., Hunt, J.F., Tong, L.To be published.