Structural basis of coagulation factor V recognition for cleavage by RVV-V
Nakayama, D., Ben Ammar, Y., Miyata, T., Takeda, S.(2011) FEBS Lett 585: 3020-3025
- PubMed: 21871889
- DOI: https://doi.org/10.1016/j.febslet.2011.08.022
- Primary Citation of Related Structures:
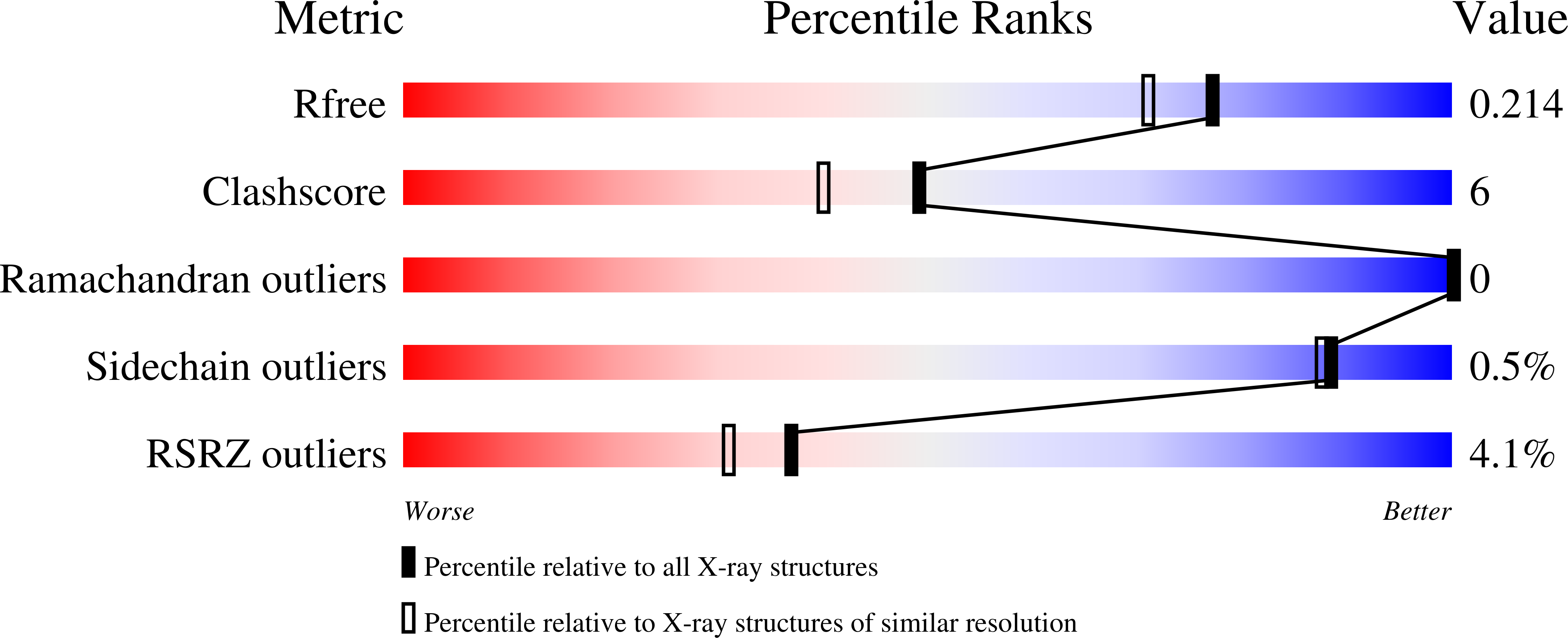
3S9A, 3S9B, 3S9C, 3SBK - PubMed Abstract:
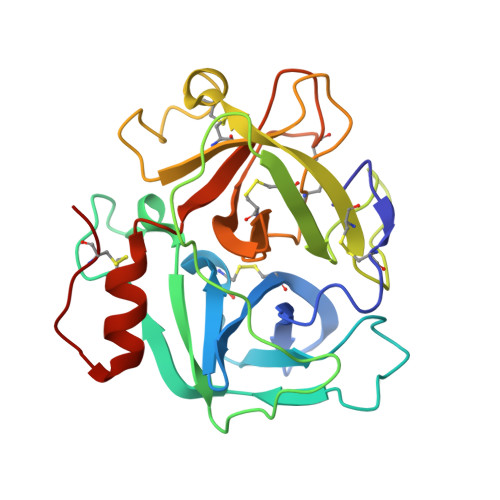
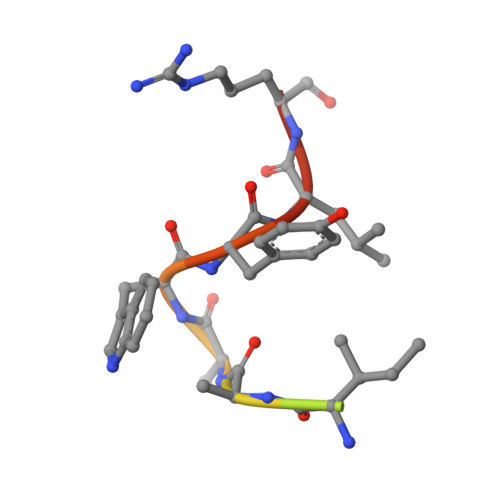
Russell's viper venom factor V (FV) activator (RVV-V) is a thrombin-like proteinase that specifically cleaves the Arg1545-Ser1546 bond of FV. Here we present the crystal structure of RVV-V in complex with the FV14 peptide (residues 1533-1546 of human FV) determined at 1.8Å resolution. The structure reveals multiple interactions between RVV-V and the seven residues, Ile1539 (P(7))-Arg1545 (P(1)), of the cleaved substrate. Comparison with substrate-free structures reveals conformational changes of the RVV-V loops upon substrate binding, suggesting that the multiple interactions are mediated by an induced-fit mechanism. The results provide an explanation for the narrow specificity of RVV-V.
Organizational Affiliation:
Department of Cardiac Physiology, National Cerebral and Cardiovascular Center Research Institute, Suita, Osaka, Japan.