Analysis of substrate specificity of Schizosaccharomyces pombe Mag1 alkylpurine DNA glycosylase.
Adhikary, S., Eichman, B.F.(2011) EMBO Rep 12: 1286-1292
- PubMed: 21960007
- DOI: https://doi.org/10.1038/embor.2011.189
- Primary Citation of Related Structures:
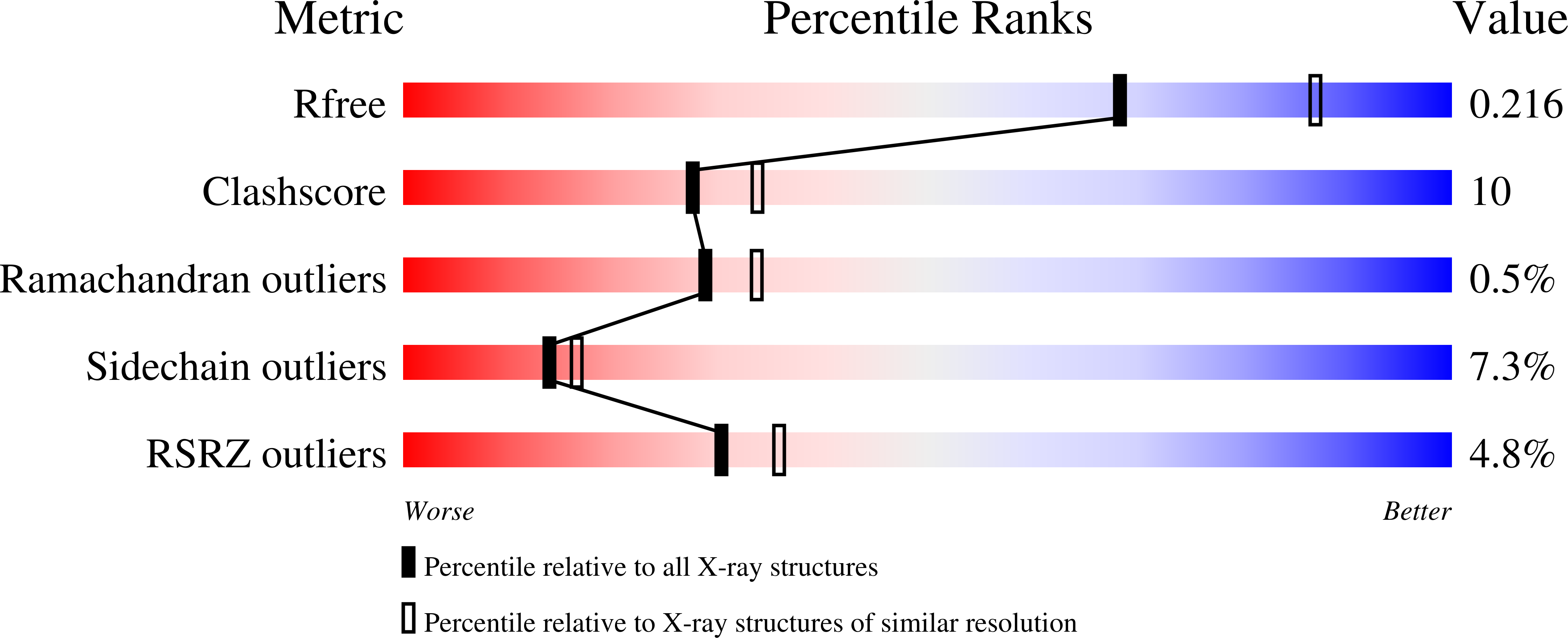
3S6I - PubMed Abstract:
DNA glycosylases specialized for the repair of alkylation damage must identify, with fine specificity, a diverse array of subtle modifications within DNA. The current mechanism involves damage sensing through interrogation of the DNA duplex, followed by more specific recognition of the target base inside the active site pocket. To better understand the physical basis for alkylpurine detection, we determined the crystal structure of Schizosaccharomyces pombe Mag1 (spMag1) in complex with DNA and performed a mutational analysis of spMag1 and the close homologue from Saccharomyces cerevisiae (scMag). Despite strong homology, spMag1 and scMag differ in substrate specificity and cellular alkylation sensitivity, although the enzymological basis for their functional differences is unknown. We show that Mag preference for 1,N(6)-ethenoadenine (ɛA) is influenced by a minor groove-interrogating residue more than the composition of the nucleobase-binding pocket. Exchanging this residue between Mag proteins swapped their ɛA activities, providing evidence that residues outside the extrahelical base-binding pocket have a role in identification of a particular modification in addition to sensing damage.
Organizational Affiliation:
Department of Biological Sciences and Center for Structural Biology, Vanderbilt University, 465 21st Avenue South, Box 351634 Station B, 5270A MRBIII, Nashville, Tennessee 37235, USA.