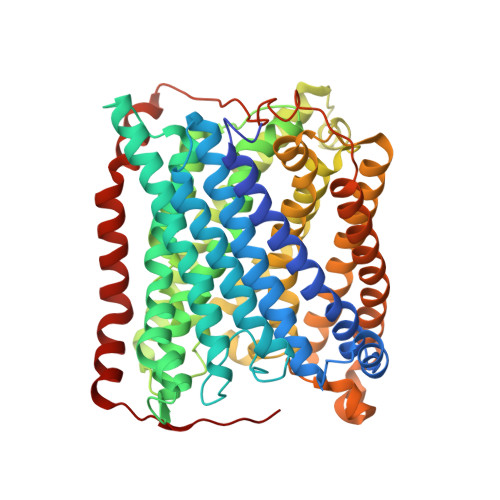
Structural changes that occur upon photolysis of the Fe(II)(a3)-CO complex in the cytochrome ba(3)-oxidase of Thermus thermophilus: A combined X-ray crystallographic and infrared spectral study demonstrates CO binding to Cu(B).
Liu, B., Zhang, Y., Sage, J.T., Soltis, S.M., Doukov, T., Chen, Y., Stout, C.D., Fee, J.A.(2012) Biochim Biophys Acta 1817: 658-665
- PubMed: 22226917
- DOI: https://doi.org/10.1016/j.bbabio.2011.12.010
- Primary Citation of Related Structures:
3QJQ, 3QJR, 3QJS, 3QJT, 3QJU, 3QJV - PubMed Abstract:
The purpose of the work was to provide a crystallographic demonstration of the venerable idea that CO photolyzed from ferrous heme-a(3) moves to the nearby cuprous ion in the cytochrome c oxidases. Crystal structures of CO-bound cytochrome ba(3)-oxidase from Thermus thermophilus, determined at ~2.8-3.2Å resolution, reveal a Fe-C distance of ~2.0Å, a Cu-O distance of 2.4Å and a Fe-C-O angle of ~126°. Upon photodissociation at 100K, X-ray structures indicate loss of Fe(a3)-CO and appearance of Cu(B)-CO having a Cu-C distance of ~1.9Å and an O-Fe distance of ~2.3Å. Absolute FTIR spectra recorded from single crystals of reduced ba(3)-CO that had not been exposed to X-ray radiation, showed several peaks around 1975cm(-1); after photolysis at 100K, the absolute FTIR spectra also showed a significant peak at 2050cm(-1). Analysis of the 'light' minus 'dark' difference spectra showed four very sharp CO stretching bands at 1970cm(-1), 1977cm(-1), 1981cm(-1), and 1985cm(-1), previously assigned to the Fe(a3)-CO complex, and a significantly broader CO stretching band centered at ~2050cm(-1), previously assigned to the CO stretching frequency of Cu(B) bound CO. As expected for light propagating along the tetragonal axis of the P4(3)2(1)2 space group, the single crystal spectra exhibit negligible dichroism. Absolute FTIR spectrometry of a CO-laden ba(3) crystal, exposed to an amount of X-ray radiation required to obtain structural data sets before FTIR characterization, showed a significant signal due to photogenerated CO(2) at 2337cm(-1) and one from traces of CO at 2133cm(-1); while bands associated with CO bound to either Fe(a3) or to Cu(B) in "light" minus "dark" FTIR difference spectra shifted and broadened in response to X-ray exposure. In spite of considerable radiation damage to the crystals, both X-ray analysis at 2.8 and 3.2Å and FTIR spectra support the long-held position that photolysis of Fe(a3)-CO in cytochrome c oxidases leads to significant trapping of the CO on the Cu(B) atom; Fe(a3) and Cu(B) ligation, at the resolutions reported here, are otherwise unaltered.
Organizational Affiliation:
The Scripps Research Institute, Department of Molecular Biology, La Jolla, CA 92037, USA.