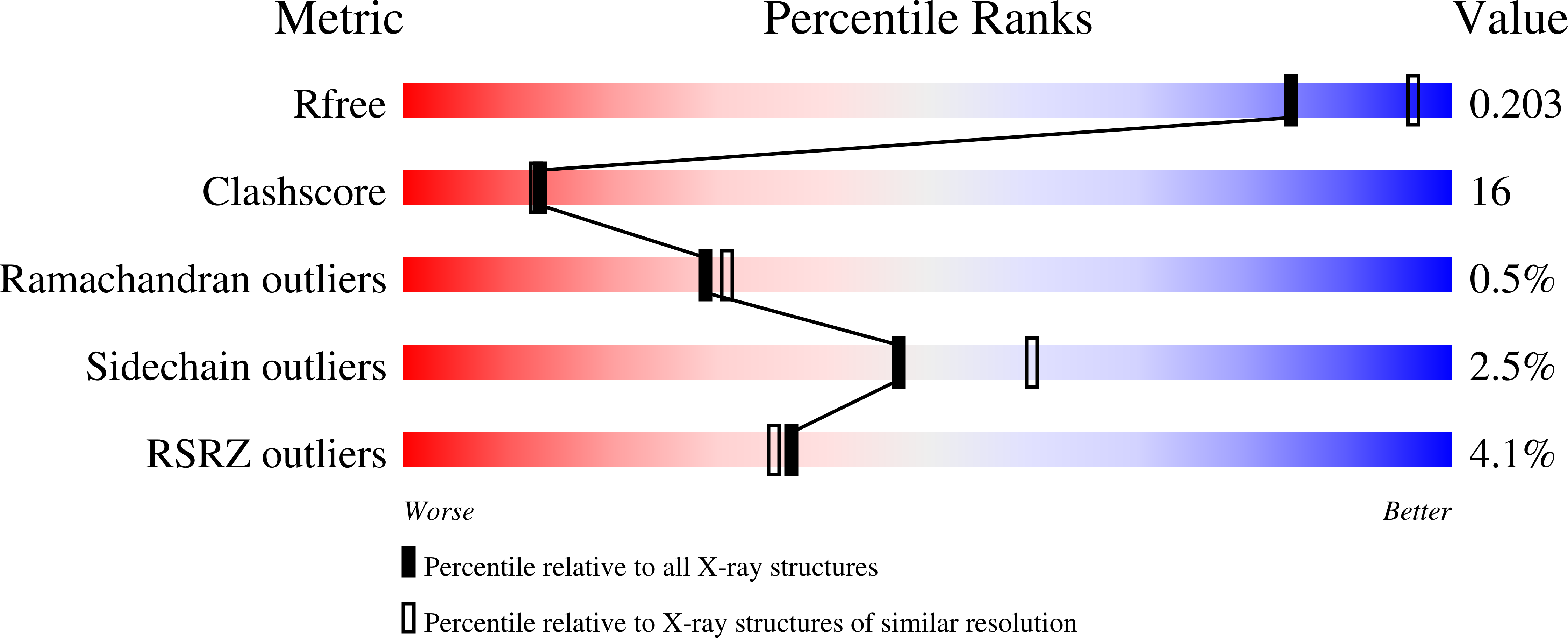
The structure of a shellfish specific GST class glutathione S-transferase from antarctic bivalve Laternula elliptica reveals novel active site architecture.
Park, A.K., Moon, J.H., Jang, E.H., Park, H., Ahn, I.Y., Lee, K.S., Chi, Y.M.(2013) Proteins 81: 531-537
- PubMed: 23152139
- DOI: https://doi.org/10.1002/prot.24208
- Primary Citation of Related Structures:
3QAV, 3QAW - PubMed Abstract:
Glutathione-S-transferases have been identified in all the living species examined so far, yet little is known about their function in marine organisms. In a previous report, the recently identified GST from Antarctic bivalve Laternula elliptica (LeGST) was classified into the rho class GST, but there are several unique features of LeGST that may justify reclassification, which could represent specific shellfish GSTs. Here, we determined the crystal structure of LeGST, which is a shellfish specific class of GST. The structural analysis showed that the relatively open and wide hydrophobic H-site of the LeGST allows this GST to accommodate various substrates. These results suggest that the H-site of LeGST may be the result of adaptation to their environments as sedentary organisms.
Organizational Affiliation:
Division of Biotechnology, College of Life Sciences, Korea University, Seoul 136-713, Republic of Korea.