The Nudix Hydrolase CDP-Chase, a CDP-Choline Pyrophosphatase, Is an Asymmetric Dimer with Two Distinct Enzymatic Activities.
Duong-Ly, K.C., Gabelli, S.B., Xu, W., Dunn, C.A., Schoeffield, A.J., Bessman, M.J., Amzel, L.M.(2011) J Bacteriol 193: 3175-3185
- PubMed: 21531795
- DOI: https://doi.org/10.1128/JB.00089-11
- Primary Citation of Related Structures:
3Q1P, 3Q4I - PubMed Abstract:
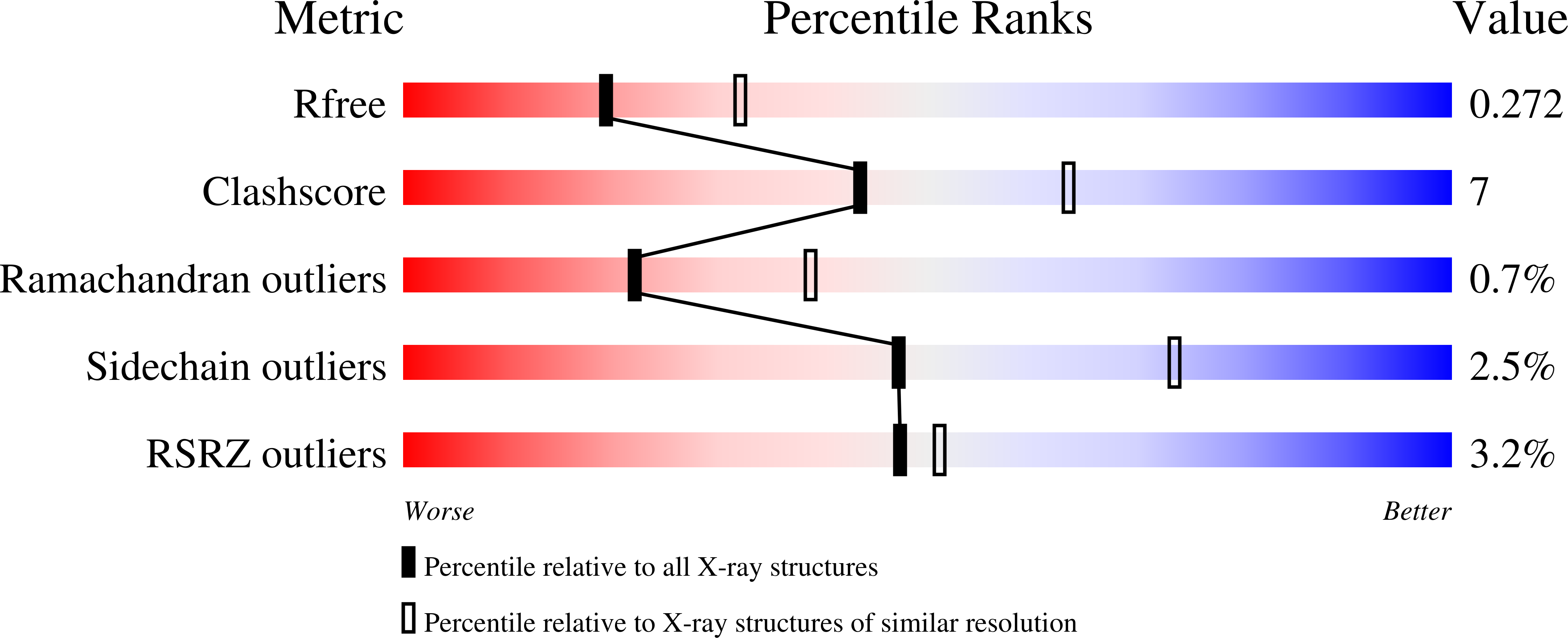
A Nudix enzyme from Bacillus cereus (NCBI RefSeq accession no. NP_831800) catalyzes the hydrolysis of CDP-choline to produce CMP and phosphocholine. Here, we show that in addition, the enzyme has a 3'→5' RNA exonuclease activity. The structure of the free enzyme, determined to a 1.8-Å resolution, shows that the enzyme is an asymmetric dimer. Each monomer consists of two domains, an N-terminal helical domain and a C-terminal Nudix domain. The N-terminal domain is placed relative to the C-terminal domain such as to result in an overall asymmetric arrangement with two distinct catalytic sites: one with an "enclosed" Nudix pyrophosphatase site and the other with a more open, less-defined cavity. Residues that may be important for determining the asymmetry are conserved among a group of uncharacterized Nudix enzymes from Gram-positive bacteria. Our data support a model where CDP-choline hydrolysis is catalyzed by the enclosed Nudix site and RNA exonuclease activity is catalyzed by the open site. CDP-Chase is the first identified member of a novel Nudix family in which structural asymmetry has a profound effect on the recognition of substrates.
Organizational Affiliation:
Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA.