Structure of ent-Copalyl Diphosphate Synthase from Arabidopsis thaliana, a Protonation-Dependent Diterpene Cyclase
Koksal, M., Hu, H., Coates, R.M., Peters, R.J., Christianson, D.W.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ent-copalyl diphosphate synthase, chloroplastic | 727 | Arabidopsis thaliana | Mutation(s): 0 Gene Names: At4g02780, GA1, T5J8.9, TPSGA1 EC: 5.5.1.13 | 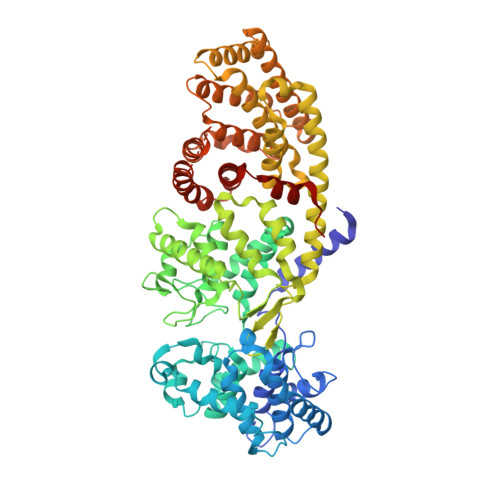 | |
UniProt | |||||
Find proteins for Q38802 (Arabidopsis thaliana) Explore Q38802 Go to UniProtKB: Q38802 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q38802 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| A3C Query on A3C | D [auth A] | 2-(methyl{2-[(1S,4aS,8aS)-5,5,8a-trimethyl-2-methylidenedecahydronaphthalen-1-yl]ethyl}amino)ethyl trihydrogen diphosphate C19 H37 N O7 P2 ZYKKZQANNLZNRN-JENIJYKNSA-N |  | ||
| 1PE Query on 1PE | E [auth A] F [auth A] G [auth A] H [auth A] I [auth B] | PENTAETHYLENE GLYCOL C10 H22 O6 JLFNLZLINWHATN-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 63.738 | α = 90 |
| b = 188.554 | β = 90 |
| c = 229.071 | γ = 90 |
| Software Name | Purpose |
|---|---|
| APS | data collection |
| PHENIX | model building |
| PHENIX | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| PHENIX | phasing |