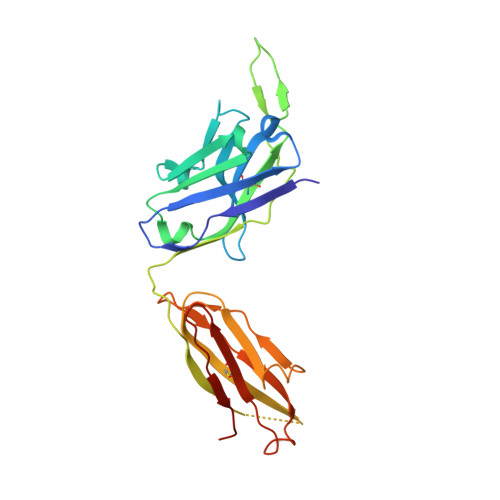
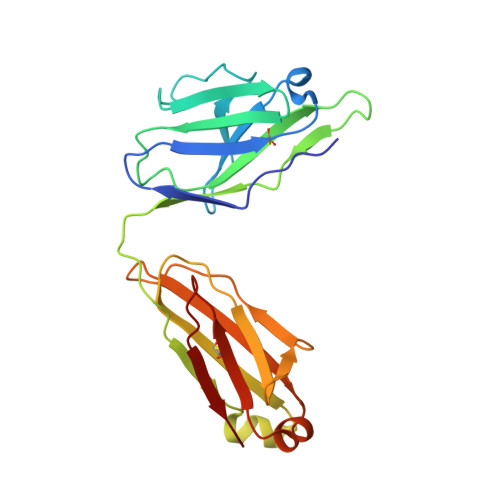
Crystal Structure of Human Antibody 2909 Reveals Conserved Features of Quaternary Structure-Specific Antibodies That Potently Neutralize HIV-1.
Changela, A., Wu, X., Yang, Y., Zhang, B., Zhu, J., Nardone, G.A., O'Dell, S., Pancera, M., Gorny, M.K., Phogat, S., Robinson, J.E., Stamatatos, L., Zolla-Pazner, S., Mascola, J.R., Kwong, P.D.(2011) J Virol 85: 2524-2535
- PubMed: 21191009
- DOI: https://doi.org/10.1128/JVI.02335-10
- Primary Citation of Related Structures:
3PIQ - PubMed Abstract:
Monoclonal antibody 2909 belongs to a class of potently neutralizing antibodies that recognize quaternary epitopes on HIV-1. Some members of this class, such as 2909, are strain specific, while others, such as antibody PG16, are broadly neutralizing; all, however, recognize a region on the gp120 envelope glycoprotein that includes two loops (V2 and V3) and forms appropriately only in the oligomeric HIV-1 spike (gp120(3)/gp41(3)). Here we present the crystal structure of 2909 and report structure-function analysis with antibody chimeras composed of 2909 and other members of this antibody class. The 2909 structure was dominated by a heavy-chain third-complementarity-determining region (CDR H3) of 21 residues, which comprised 36% of the combining surface and formed a β-hairpin club extending ∼20 Å beyond the rest of the antibody. Sequence analysis and mass spectrometry identified sites of tyrosine sulfation at the middle and top of CDR H3; substitutions with phenylalanine either ablated (middle substitution) or substantially diminished (top substitution) neutralization. Chimeric antibodies composed of heavy and light chains, exchanged between 2909 and other members of the class, indicated a substantial lack of complementation. Comparison of 2909 to PG16 (which is tyrosine sulfated and the only other member of the class for which a structure has previously been reported) showed that both utilize protruding, anionic CDR H3s for recognition. Thus, despite some diversity, members of this class share structural and functional similarities, with conserved features of the CDR H3 subdomain likely reflecting prevalent solutions by the human immune system for recognition of a quaternary site of HIV-1 vulnerability.
Organizational Affiliation:
Vaccine Research Center, NIAID/NIH, 40 Convent Drive, Building 40, Room 4508, Bethesda, MD 20892-3027, USA.