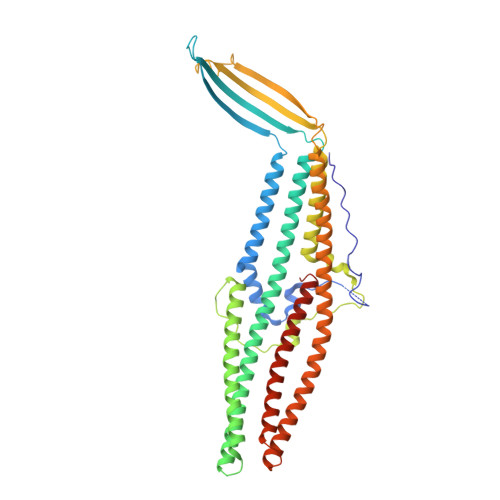
Crystal Structure of Escherichia coli CusC, the Outer Membrane Component of a Heavy Metal Efflux Pump.
Kulathila, R., Kulathila, R., Indic, M., van den Berg, B.(2011) PLoS One 6: e15610-e15610
- PubMed: 21249122
- DOI: https://doi.org/10.1371/journal.pone.0015610
- Primary Citation of Related Structures:
3PIK - PubMed Abstract:
While copper has essential functions as an enzymatic co-factor, excess copper ions are toxic for cells, necessitating mechanisms for regulating its levels. The cusCBFA operon of E. coli encodes a four-component efflux pump dedicated to the extrusion of Cu(I) and Ag(I) ions. We have solved the X-ray crystal structure of CusC, the outer membrane component of the Cus heavy metal efflux pump, to 2.3 Å resolution. The structure has the largest extracellular opening of any outer membrane factor (OMF) protein and suggests, for the first time, the presence of a tri-acylated N-terminal lipid anchor. The CusC protein does not have any obvious features that would make it specific for metal ions, suggesting that the narrow substrate specificity of the pump is provided by other components of the pump, most likely by the inner membrane component CusA.
Organizational Affiliation:
University of Massachusetts Medical School, Program in Molecular Medicine, Worcester, Massachusetts, United States of America.