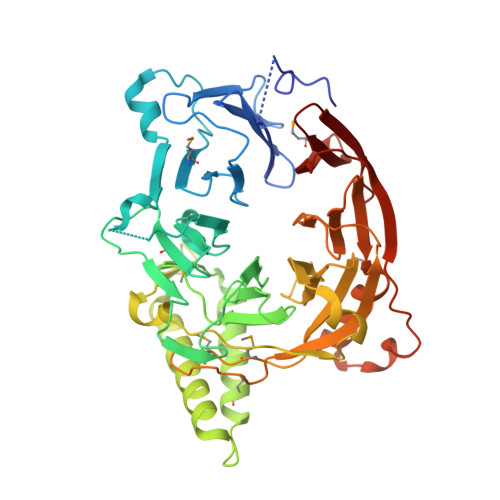
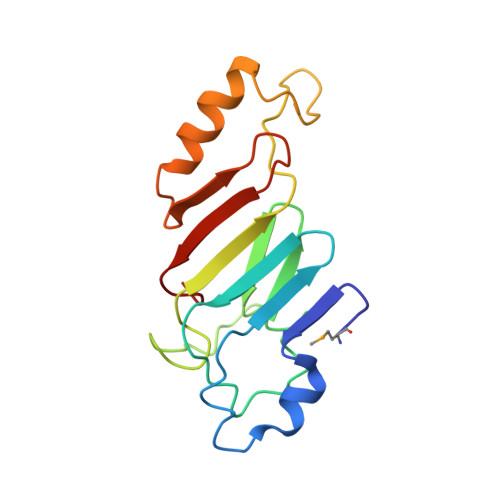
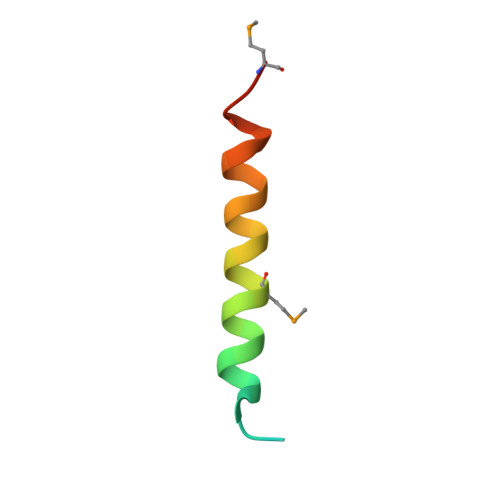
Structural and functional analysis of an essential nucleoporin heterotrimer on the cytoplasmic face of the nuclear pore complex.
Yoshida, K., Seo, H.S., Debler, E.W., Blobel, G., Hoelz, A.(2011) Proc Natl Acad Sci U S A 108: 16571-16576
- PubMed: 21930948
- DOI: https://doi.org/10.1073/pnas.1112846108
- Primary Citation of Related Structures:
3PBP - PubMed Abstract:
So far, only a few of the interactions between the ≈ 30 nucleoporins comprising the modular structure of the nuclear pore complex have been defined at atomic resolution. Here we report the crystal structure, at 2.6 Å resolution, of a heterotrimeric complex, composed of fragments of three cytoplasmically oriented nucleoporins of yeast: Nup82, Nup116, and Nup159. Our data show that the Nup82 fragment, representing more than the N-terminal half of the molecule, folds into an extensively decorated, seven-bladed β-propeller that forms the centerpiece of this heterotrimeric complex and anchors both a C-terminal fragment of Nup116 and the C-terminal tail of Nup159. Binding between Nup116 and Nup82 is mutually reinforced via two loops, one emanating from the Nup82 β-propeller and the other one from the β-sandwich fold of Nup116, each contacting binding pockets in their counterparts. The Nup82-Nup159 interaction occurs through an amphipathic α-helix of Nup159, which is cradled in a large hydrophobic groove that is generated from several large surface decorations of the Nup82 β-propeller. Although Nup159 and Nup116 fragments bind to the Nup82 β-propeller in close vicinity, there are no direct contacts between them, consistent with the noncooperative binding that was detected biochemically. Extensive mutagenesis delineated hot-spot residues for these interactions. We also showed that the Nup82 β-propeller binds to other yeast Nup116 family members, Nup145N, Nup100 and to the mammalian homolog, Nup98. Notably, each of the three nucleoporins contains additional nuclear pore complex binding sites, distinct from those that were defined here in the heterotrimeric Nup82•Nup159•Nup116 complex.
Organizational Affiliation:
Laboratory of Cell Biology, Howard Hughes Medical Institute, The Rockefeller University, 1230 York Avenue, New York, NY 10065, USA.