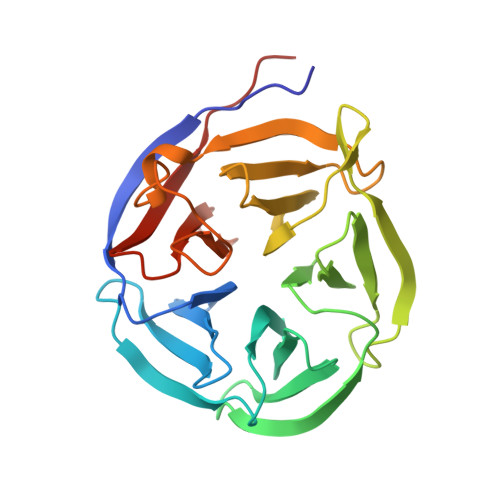
Crystal structure and functional analysis of the glutaminyl cyclase from Xanthomonas campestris
Huang, W.-L., Wang, Y.-R., Ko, T.-P., Chia, C.-Y., Huang, K.-F., Wang, A.H.-J.(2010) J Mol Biol 401: 374-388
- PubMed: 20558177
- DOI: https://doi.org/10.1016/j.jmb.2010.06.012
- Primary Citation of Related Structures:
3MBR - PubMed Abstract:
Glutaminyl cyclases (QCs) (EC 2.3.2.5) catalyze the formation of pyroglutamate (pGlu) at the N-terminus of many proteins and peptides, a critical step for the maturation of these bioactive molecules. Proteins having QC activity have been identified in animals and plants, but not in bacteria. Here, we report the first bacterial QC from the plant pathogen Xanthomonas campestris (Xc). The crystal structure of the enzyme was solved and refined to 1.44-A resolution. The structure shows a five-bladed beta-propeller and exhibits a scaffold similar to that of papaya QC (pQC), but with some sequence deletions and conformational changes. In contrast to the pQC structure, the active site of XcQC has a wider substrate-binding pocket, but its accessibility is modulated by a protruding loop acting as a flap. Enzyme activity analyses showed that the wild-type XcQC possesses only 3% QC activity compared to that of pQC. Superposition of those two structures revealed that an active-site glutamine residue in pQC is substituted by a glutamate (Glu(45)) in XcQC, although position 45 is a glutamine in most bacterial QC sequences. The E45Q mutation increased the QC activity by an order of magnitude, but the mutation E45A led to a drop in the enzyme activity, indicating the critical catalytic role of this residue. Further mutagenesis studies support the catalytic role of Glu(89) as proposed previously and confirm the importance of several conserved amino acids around the substrate-binding pocket. XcQC was shown to be weakly resistant to guanidine hydrochloride, extreme pH, and heat denaturations, in contrast to the extremely high stability of pQC, despite their similar scaffold. On the basis of structure comparison, the low stability of XcQC may be attributed to the absence of both a disulfide linkage and some hydrogen bonds in the closure of beta-propeller structure. These results significantly improve our understanding of the catalytic mechanism and extreme stability of type I QCs, which will be useful in further applications of QC enzymes.
Organizational Affiliation:
Graduate Institute of Biochemical Science, College of Life Science, National Taiwan University, Taipei 106, Taiwan.