Inhibition and Structure of Toxoplasma gondii Purine Nucleoside Phosphorylase.
Donaldson, T.M., Cassera, M.B., Ho, M.C., Zhan, C., Merino, E.F., Evans, G.B., Tyler, P.C., Almo, S.C., Schramm, V.L., Kim, K.(2014) Eukaryot Cell 13: 572-579
- PubMed: 24585883
- DOI: https://doi.org/10.1128/EC.00308-13
- Primary Citation of Related Structures:
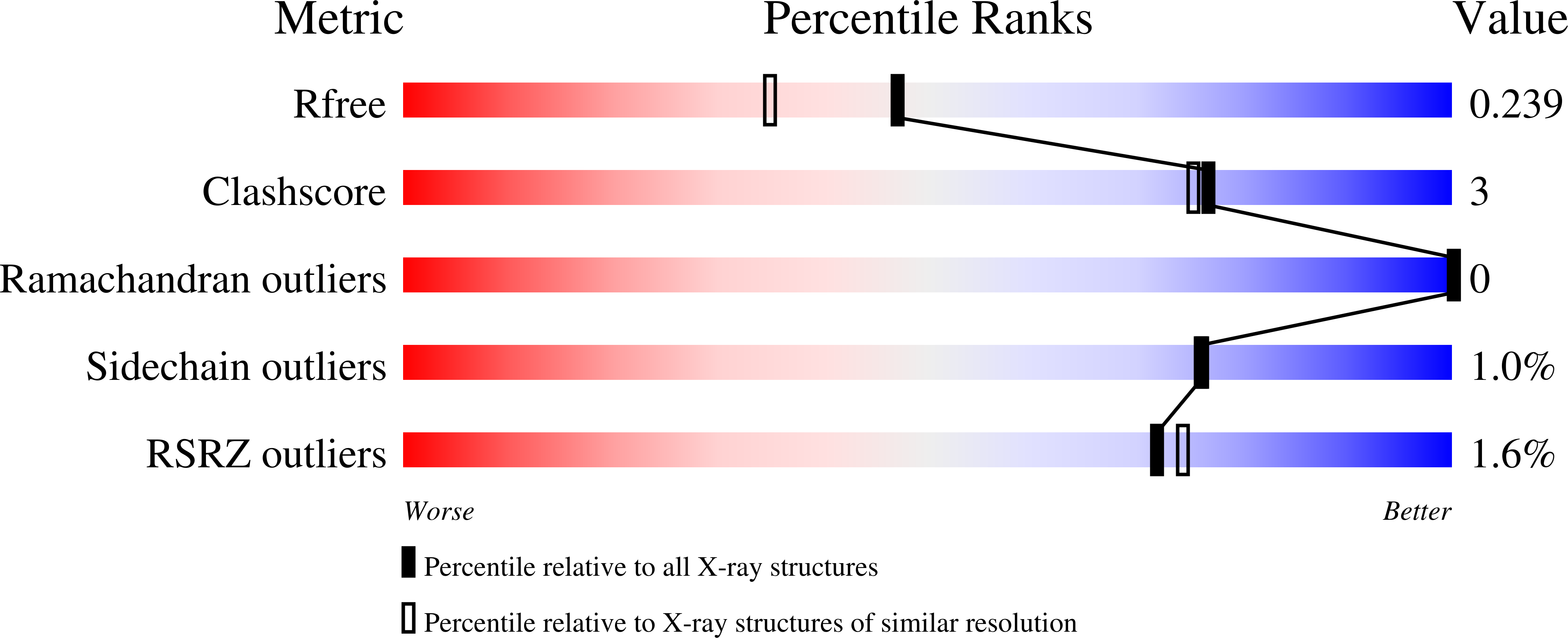
3MB8 - PubMed Abstract:
The intracellular pathogen Toxoplasma gondii is a purine auxotroph that relies on purine salvage for proliferation. We have optimized T. gondii purine nucleoside phosphorylase (TgPNP) stability and crystallized TgPNP with phosphate and immucillin-H, a transition-state analogue that has high affinity for the enzyme. Immucillin-H bound to TgPNP with a dissociation constant of 370 pM, the highest affinity of 11 immucillins selected to probe the catalytic site. The specificity for transition-state analogues indicated an early dissociative transition state for TgPNP. Compared to Plasmodium falciparum PNP, large substituents surrounding the 5'-hydroxyl group of inhibitors demonstrate reduced capacity for TgPNP inhibition. Catalytic discrimination against large 5' groups is consistent with the inability of TgPNP to catalyze the phosphorolysis of 5'-methylthioinosine to hypoxanthine. In contrast to mammalian PNP, the 2'-hydroxyl group is crucial for inhibitor binding in the catalytic site of TgPNP. This first crystal structure of TgPNP describes the basis for discrimination against 5'-methylthioinosine and similarly 5'-hydroxy-substituted immucillins; structural differences reflect the unique adaptations of purine salvage pathways of Apicomplexa.
Organizational Affiliation:
Department of Microbiology and Immunology, Albert Einstein College of Medicine, Yeshiva University, Bronx, New York, USA.