Structural characterization of acylimine-containing blue and red chromophores in mTagBFP and TagRFP fluorescent proteins.
Subach, O.M., Malashkevich, V.N., Zencheck, W.D., Morozova, K.S., Piatkevich, K.D., Almo, S.C., Verkhusha, V.V.(2010) Chem Biol 17: 333-341
- PubMed: 20416505
- DOI: https://doi.org/10.1016/j.chembiol.2010.03.005
- Primary Citation of Related Structures:
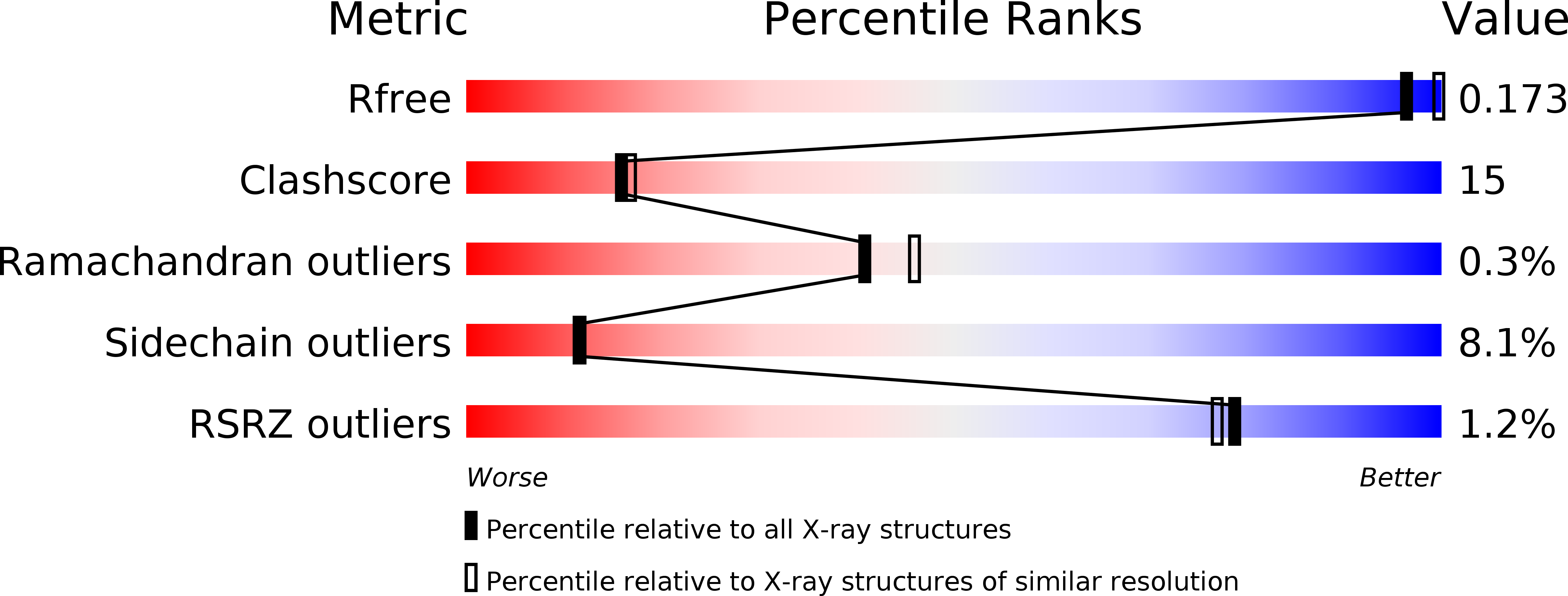
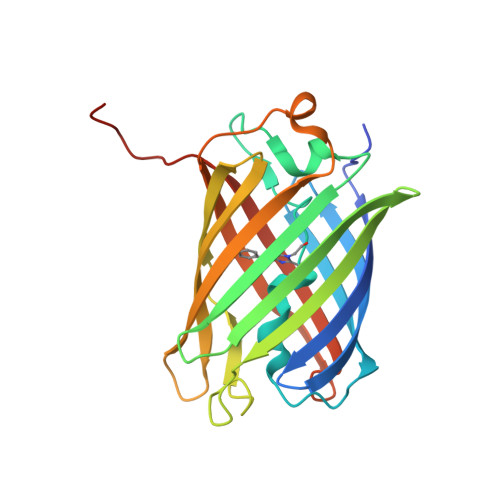
3M22, 3M24 - PubMed Abstract:
We determined the 2.2 A crystal structures of the red fluorescent protein TagRFP and its derivative, the blue fluorescent protein mTagBFP. The crystallographic analysis is consistent with a model in which TagRFP has the trans coplanar anionic chromophore with the conjugated pi-electron system, similar to that of DsRed-like chromophores. Refined conformation of mTagBFP suggests the presence of an N-acylimine functionality in its chromophore and single C(alpha)-C(beta) bond in the Tyr64 side chain. Mass spectrum of mTagBFP chromophore-bearing peptide indicates a loss of 20 Da upon maturation, whereas tandem mass spectrometry reveals that the C(alpha)-N bond in Leu63 is oxidized. These data indicate that mTagBFP has a new type of the chromophore, N-[(5-hydroxy-1H-imidazole-2-yl)methylidene]acetamide. We propose a chemical mechanism in which the DsRed-like chromophore is formed via the mTagBFP-like blue intermediate.
Organizational Affiliation:
Department of Anatomy and Structural Biology and Gruss-Lipper Biophotonics Center, Albert Einstein College of Medicine, 1300 Morris Park Avenue, Bronx, NY10461, USA.