Holo-Ni(II)HpNikR Is an Asymmetric Tetramer Containing Two Different Nickel-Binding Sites.
West, A.L., St John, F., Lopes, P.E., Mackerell, A.D., Pozharski, E., Michel, S.L.(2010) J Am Chem Soc 132: 14447-14456
- PubMed: 20863122
- DOI: https://doi.org/10.1021/ja104118r
- Primary Citation of Related Structures:
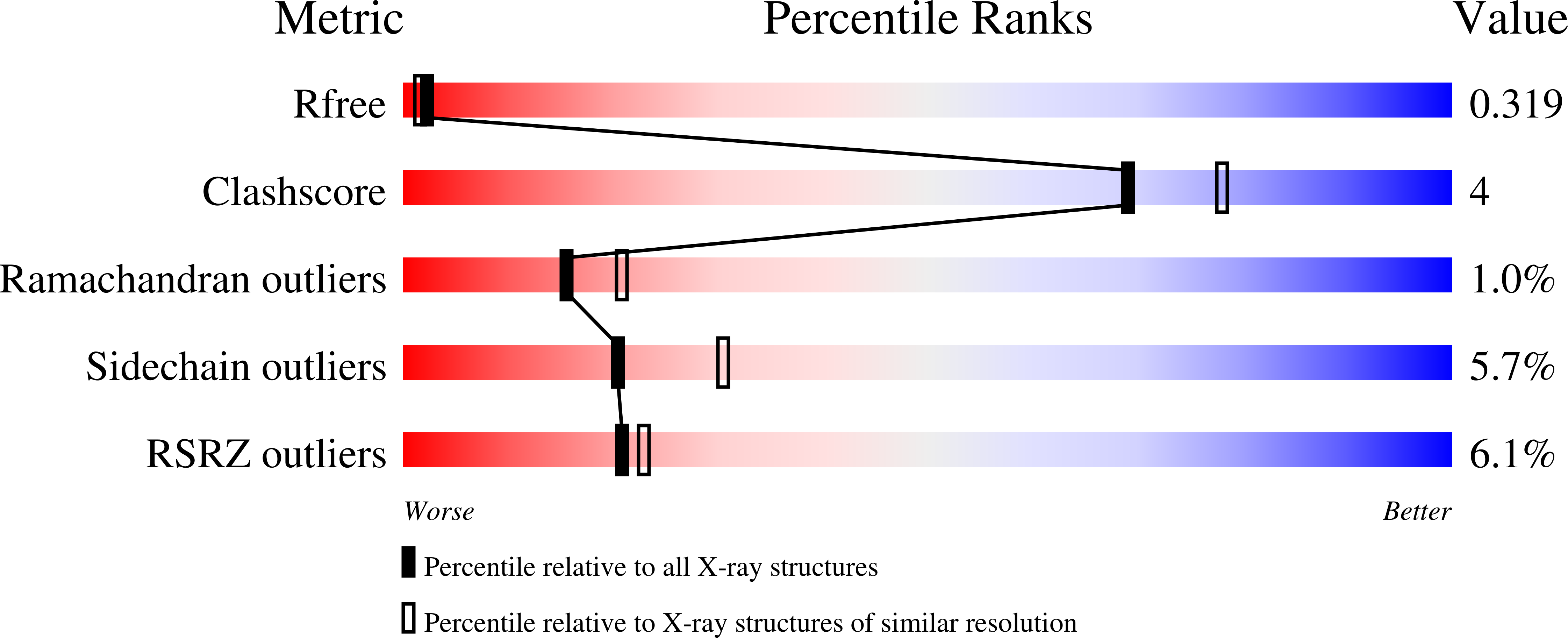
3LGH - PubMed Abstract:
The metalloregulatory protein NikR from Helicobacter pylori (HpNikR) is a master regulator of gene expression which both activates and represses specific genes in response to nickel availability. Here, we report the first crystal structure (at 2.37 Å resolution) of Ni(II)HpNikR prepared directly from the holo protein. The protein contains four nickel ions located in two distinct coordination environments. Two nickel ions are bound to sites in a four-coordinate square-planar geometry as predicted on the basis of the structures of NikR from Escherichia coli and Pyrococcus horikoshii . The remaining two nickel ions are bound to sites with unexpected 5- or 6-coordination geometries which were previously thought to be involved in nickel incorporation into the protein. The nickel with 5-/6-coordination geometry utilizes three histidines from two separate monomeric HpNikR units along with two or three water molecules as ligands. The spatial location of the nickel in the 5-/6-coordinate site is within approximately 5 Å of the expected site if a 4-coordinate square-planar geometry occurred. Two of the histidines that participate as ligands in the 5-/6-coordinate site would also participate as ligands if the 4-coordinate site was occupied, making it impossible for both sites to be occupied simultaneously. DFT calculations show that the 5-/6-coordinate geometries are energetically favorable when the local protein environment is included in the calculations. The presence of two distinct coordination environments in HpNikR is suggested to be related to the specificity and binding affinity of this transcription factor for DNA.
Organizational Affiliation:
Department of Pharmaceutical Sciences, School of Pharmacy, University of Maryland, Baltimore, Maryland 21201-1180, USA.