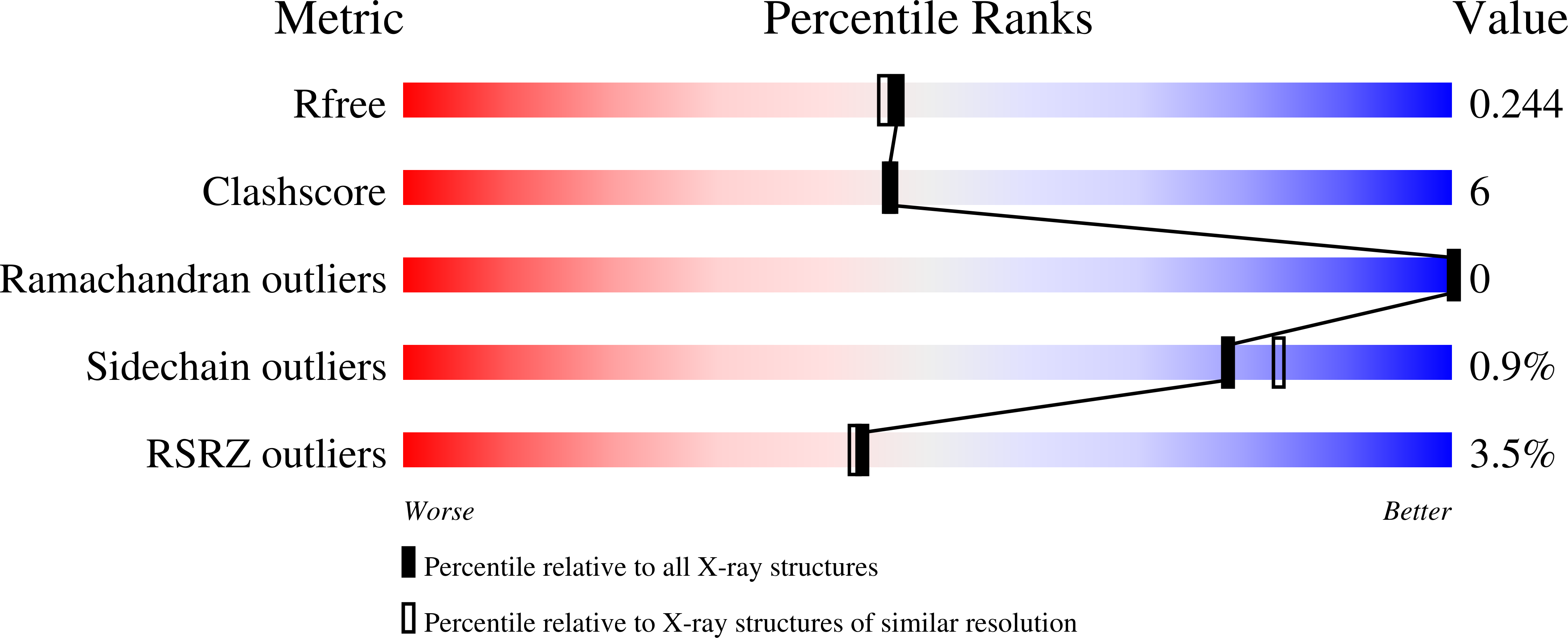
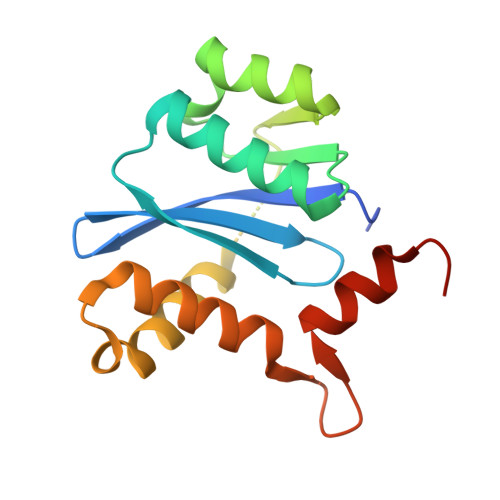
Crystal structure of the HIV-1 integrase core domain in complex with sucrose reveals details of an allosteric inhibitory binding site
Wielens, J., Headey, S.J., Jeevarajah, D., Rhodes, D.I., Deadman, J., Chalmers, D.K., Scanlon, M.J., Parker, M.W.(2010) FEBS Lett 584: 1455-1462
- PubMed: 20227411
- DOI: https://doi.org/10.1016/j.febslet.2010.03.016
- Primary Citation of Related Structures:
3L3U, 3L3V - PubMed Abstract:
HIV integrase (IN) is an essential enzyme in HIV replication and an important target for drug design. IN has been shown to interact with a number of cellular and viral proteins during the integration process. Disruption of these important interactions could provide a mechanism for allosteric inhibition of IN. We present the highest resolution crystal structure of the IN core domain to date. We also present a crystal structure of the IN core domain in complex with sucrose which is bound at the dimer interface in a region that has previously been reported to bind integrase inhibitors.
Organizational Affiliation:
Medicinal Chemistry and Drug Action, Monash Institute of Pharmaceutical Sciences, Parkville, Victoria, Australia.