T cell allorecognition via molecular mimicry.
Macdonald, W.A., Chen, Z., Gras, S., Archbold, J.K., Tynan, F.E., Clements, C.S., Bharadwaj, M., Kjer-Nielsen, L., Saunders, P.M., Wilce, M.C., Crawford, F., Stadinsky, B., Jackson, D., Brooks, A.G., Purcell, A.W., Kappler, J.W., Burrows, S.R., Rossjohn, J., McCluskey, J.(2009) Immunity 31: 897-908
- PubMed: 20064448
- DOI: https://doi.org/10.1016/j.immuni.2009.09.025
- Primary Citation of Related Structures:
3KPL, 3KPM, 3KPN, 3KPO, 3KPP, 3KPQ, 3KPR, 3KPS - PubMed Abstract:
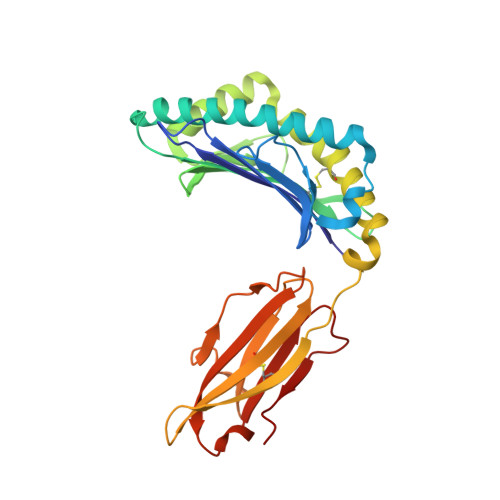
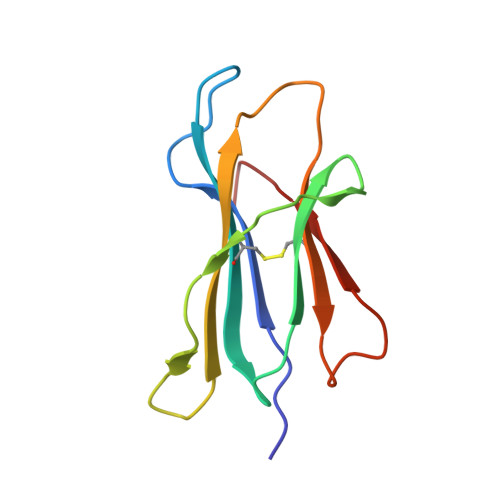
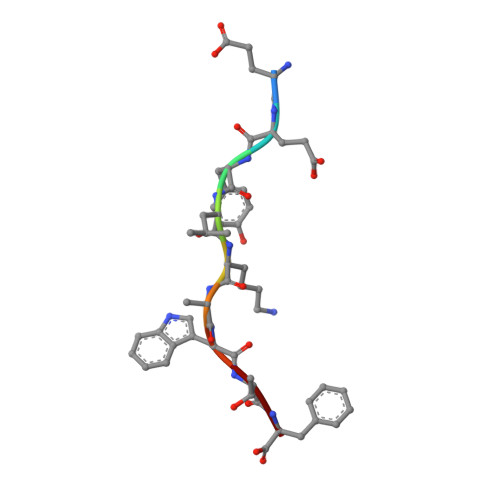
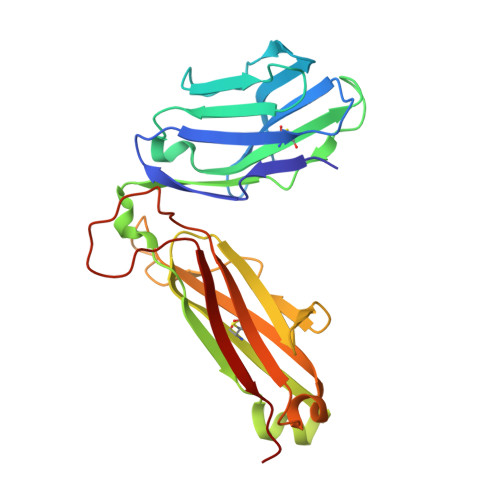
T cells often alloreact with foreign human leukocyte antigens (HLA). Here we showed the LC13 T cell receptor (TCR), selected for recognition on self-HLA-B( *)0801 bound to a viral peptide, alloreacts with B44 allotypes (HLA-B( *)4402 and HLA-B( *)4405) bound to two different allopeptides. Despite extensive polymorphism between HLA-B( *)0801, HLA-B( *)4402, and HLA-B( *)4405 and the disparate sequences of the viral and allopeptides, the LC13 TCR engaged these peptide-HLA (pHLA) complexes identically, accommodating mimicry of the viral peptide by the allopeptide. The viral and allopeptides adopted similar conformations only after TCR ligation, revealing an induced-fit mechanism of molecular mimicry. The LC13 T cells did not alloreact against HLA-B( *)4403, and the single residue polymorphism between HLA-B( *)4402 and HLA-B( *)4403 affected the plasticity of the allopeptide, revealing that molecular mimicry was associated with TCR specificity. Accordingly, molecular mimicry that is HLA and peptide dependent is a mechanism for human T cell alloreactivity between disparate cognate and allogeneic pHLA complexes.
Organizational Affiliation:
The Protein Crystallography Unit, Department of Biochemistry and Molecular Biology, Monash University, Clayton, Victoria 3800, Australia.