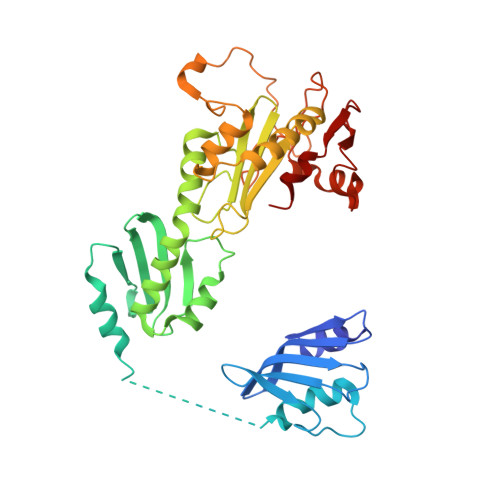
The structure of the catalytic subunit FANCL of the Fanconi anemia core complex
Cole, A.R., Lewis, L.P.C., Walden, H.(2010) Nat Struct Mol Biol 17: 294-298
- PubMed: 20154706
- DOI: https://doi.org/10.1038/nsmb.1759
- Primary Citation of Related Structures:
3K1L - PubMed Abstract:
The Fanconi anemia (FA) pathway is activated in response to DNA damage, leading to monoubiquitination of the substrates FANCI and FANCD2 by the FA core complex. Here we report the crystal structure of FANCL, the catalytic subunit of the FA core complex, at 3.2 A. The structure reveals an architecture fundamentally different from previous sequence-based predictions. The molecule is composed of an N-terminal E2-like fold, which we term the ELF domain, a novel double-RWD (DRWD) domain, and a C-terminal really interesting new gene (RING) domain predicted to facilitate E2 binding. Binding assays show that the DRWD domain, but not the ELF domain, is responsible for substrate binding.
Organizational Affiliation:
Protein Structure and Function Laboratory, Lincoln's Inn Fields Laboratories of the London Research Institute, Cancer Research UK, London Research Institute, London, UK.