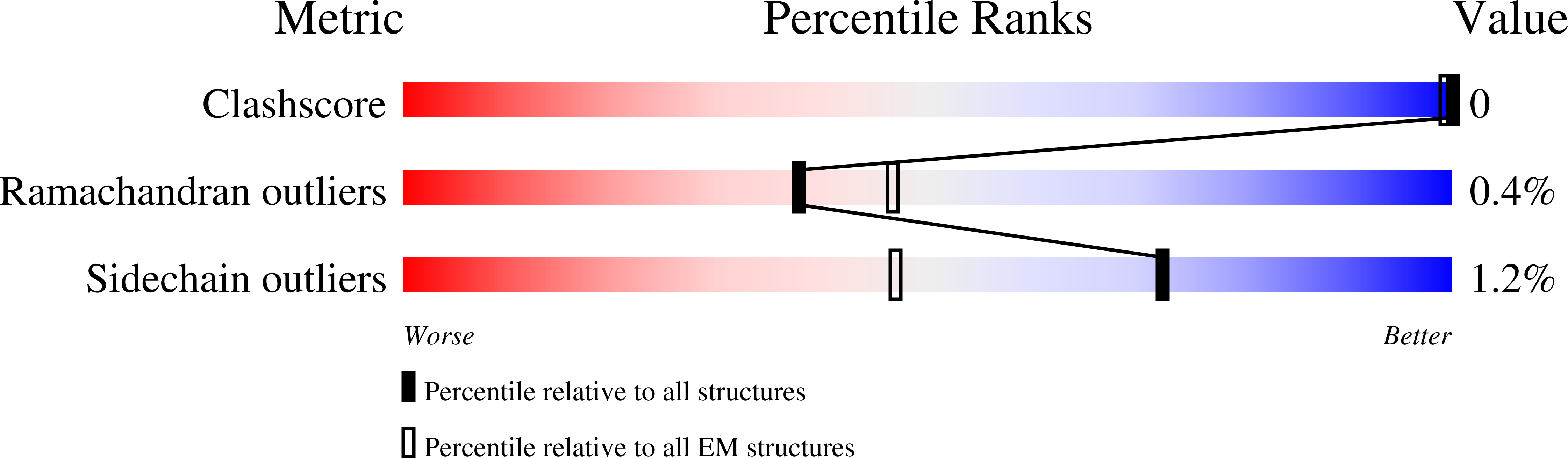CryoEM and computer simulations reveal a novel kinase conformational switch in bacterial chemotaxis signaling.
Cassidy, C.K., Himes, B.A., Alvarez, F.J., Ma, J., Zhao, G., Perilla, J.R., Schulten, K., Zhang, P.(2015) Elife 4: e08419-e08419
- PubMed: 26583751
- DOI: https://doi.org/10.7554/eLife.08419
- Primary Citation of Related Structures:
3JA6 - PubMed Abstract:
Chemotactic responses in bacteria require large, highly ordered arrays of sensory proteins to mediate the signal transduction that ultimately controls cell motility. A mechanistic understanding of the molecular events underlying signaling, however, has been hampered by the lack of a high-resolution structural description of the extended array. Here, we report a novel reconstitution of the array, involving the receptor signaling domain, histidine kinase CheA, and adaptor protein CheW, as well as a density map of the core-signaling unit at 11.3 Å resolution, obtained by cryo-electron tomography and sub-tomogram averaging. Extracting key structural constraints from our density map, we computationally construct and refine an atomic model of the core array structure, exposing novel interfaces between the component proteins. Using all-atom molecular dynamics simulations, we further reveal a distinctive conformational change in CheA. Mutagenesis and chemical cross-linking experiments confirm the importance of the conformational dynamics of CheA for chemotactic function.
Organizational Affiliation:
Department of Physics and Beckman Institute, University of Illinois at Urbana-Champaign, Urbana, United States.