Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota.
Hehemann, J.H., Correc, G., Barbeyron, T., Helbert, W., Czjzek, M., Michel, G.(2010) Nature 464: 908-912
- PubMed: 20376150
- DOI: https://doi.org/10.1038/nature08937
- Primary Citation of Related Structures:
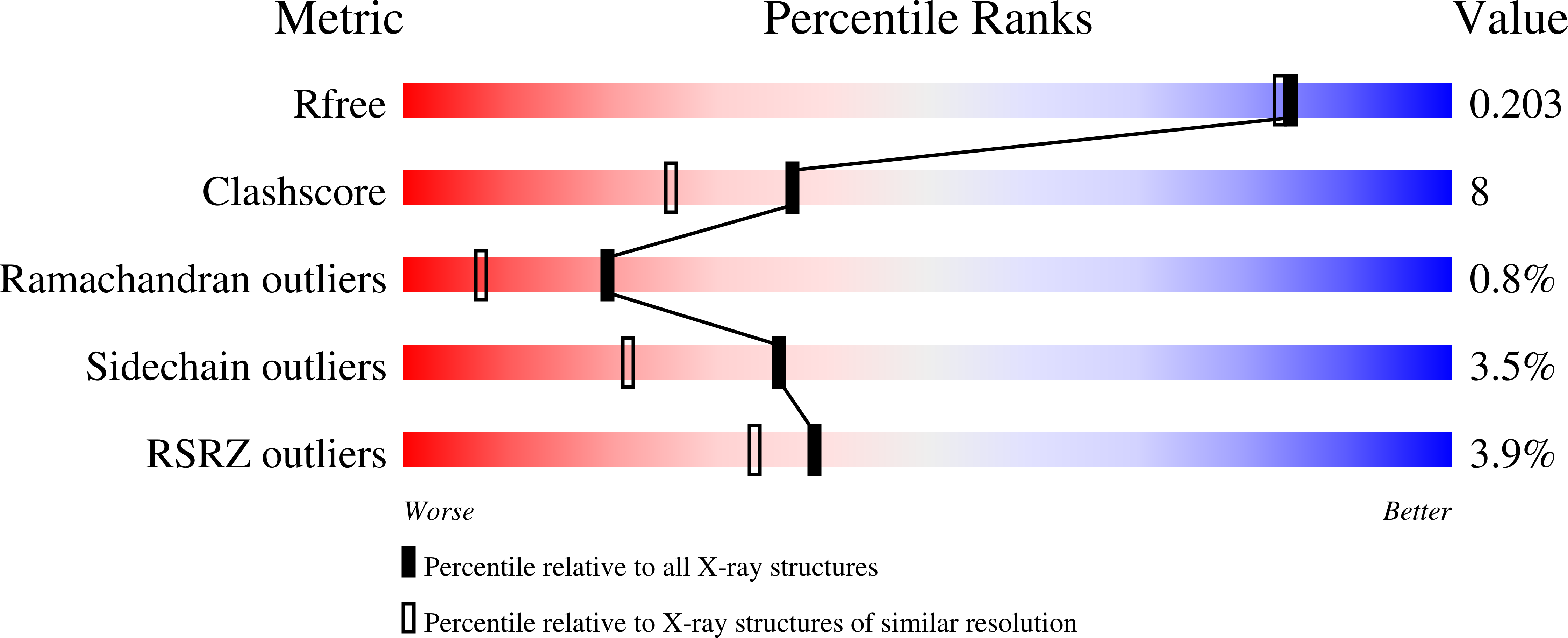
3ILF, 3JUU - PubMed Abstract:
Gut microbes supply the human body with energy from dietary polysaccharides through carbohydrate active enzymes, or CAZymes, which are absent in the human genome. These enzymes target polysaccharides from terrestrial plants that dominated diet throughout human evolution. The array of CAZymes in gut microbes is highly diverse, exemplified by the human gut symbiont Bacteroides thetaiotaomicron, which contains 261 glycoside hydrolases and polysaccharide lyases, as well as 208 homologues of susC and susD-genes coding for two outer membrane proteins involved in starch utilization. A fundamental question that, to our knowledge, has yet to be addressed is how this diversity evolved by acquiring new genes from microbes living outside the gut. Here we characterize the first porphyranases from a member of the marine Bacteroidetes, Zobellia galactanivorans, active on the sulphated polysaccharide porphyran from marine red algae of the genus Porphyra. Furthermore, we show that genes coding for these porphyranases, agarases and associated proteins have been transferred to the gut bacterium Bacteroides plebeius isolated from Japanese individuals. Our comparative gut metagenome analyses show that porphyranases and agarases are frequent in the Japanese population and that they are absent in metagenome data from North American individuals. Seaweeds make an important contribution to the daily diet in Japan (14.2 g per person per day), and Porphyra spp. (nori) is the most important nutritional seaweed, traditionally used to prepare sushi. This indicates that seaweeds with associated marine bacteria may have been the route by which these novel CAZymes were acquired in human gut bacteria, and that contact with non-sterile food may be a general factor in CAZyme diversity in human gut microbes.
Organizational Affiliation:
Université Pierre et Marie Curie, Paris, France.