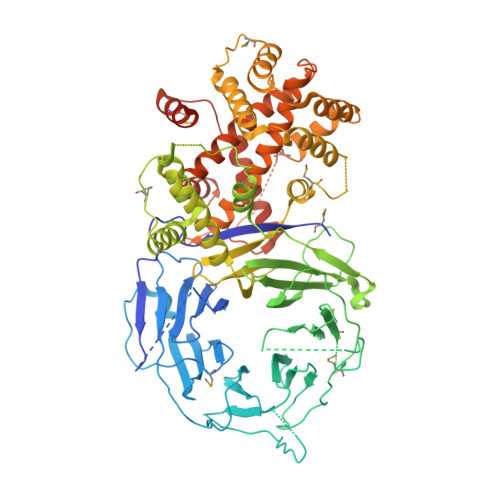
The structure of the scaffold nucleoporin Nup120 reveals a new and unexpected domain architecture.
Leksa, N.C., Brohawn, S.G., Schwartz, T.U.(2009) Structure 17: 1082-1091
- PubMed: 19576787
- DOI: https://doi.org/10.1016/j.str.2009.06.003
- Primary Citation of Related Structures:
3HXR - PubMed Abstract:
Nucleocytoplasmic transport is mediated by nuclear pore complexes (NPCs), enormous protein assemblies residing in circular openings in the nuclear envelope. The NPC is modular, with transient and stable components. The stable core is essentially built from two multiprotein complexes, the Y-shaped heptameric Nup84 complex and the Nic96 complex, arranged around an eightfold axis. We present the crystal structure of Nup120(1-757), one of the two short arms of the Y-shaped Nup84 complex. The protein adopts a compact oval shape built around a novel bipartite alpha-helical domain intimately integrated with a beta-propeller domain. The domain arrangement is substantially different from the Nup85*Seh1 complex, which forms the other short arm of the Y. With the data presented here, we establish that all three branches of the Y-shaped Nup84 complex are tightly connected by helical interactions and that the beta-propellers likely form interaction site(s) to neighboring complexes.
Organizational Affiliation:
Department of Biology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.