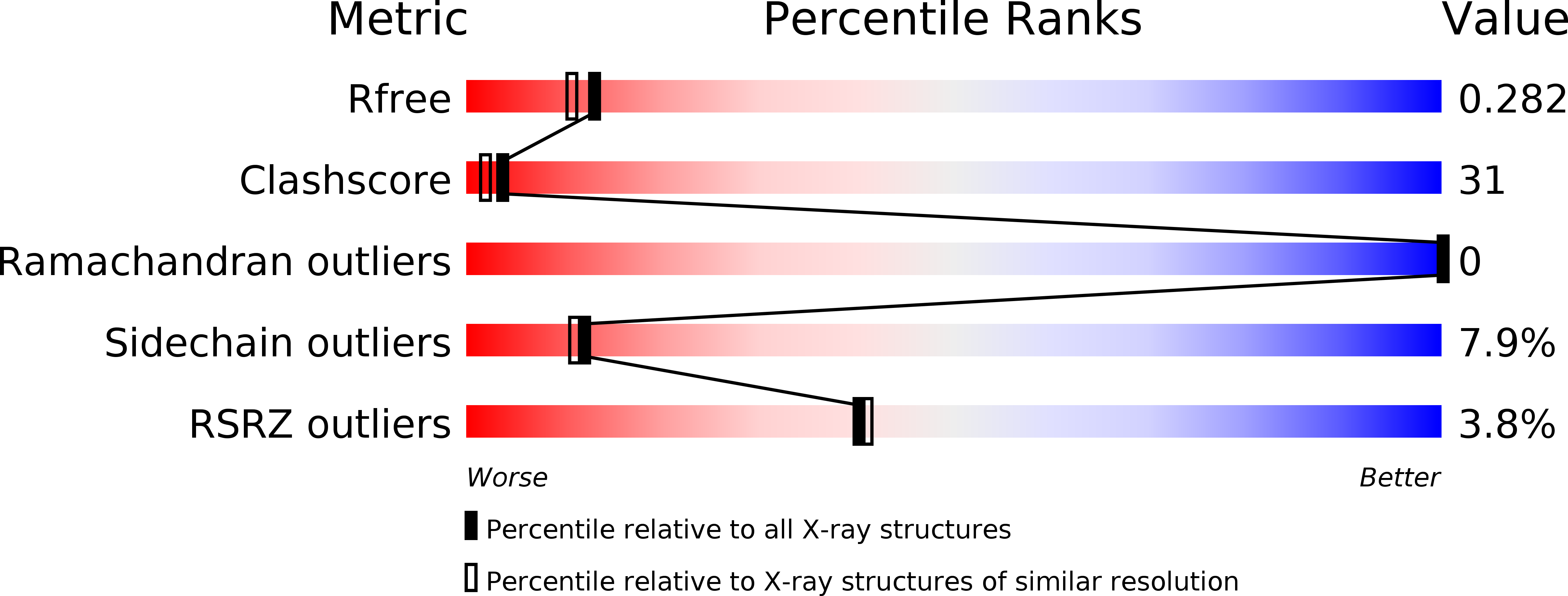
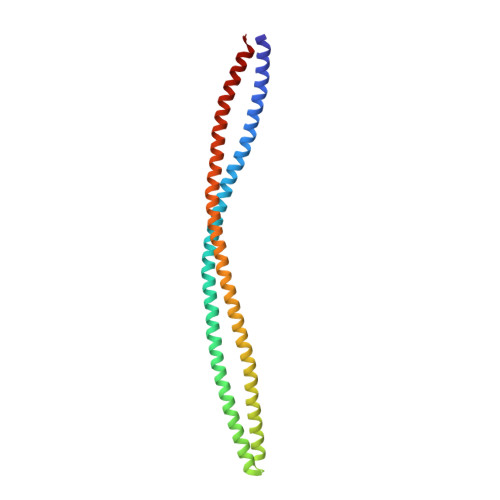The structure of a soluble chemoreceptor suggests a mechanism for propagating conformational signals.
Pollard, A.M., Bilwes, A.M., Crane, B.R.(2009) Biochemistry 48: 1936-1944
- PubMed: 19149470
- DOI: https://doi.org/10.1021/bi801727m
- Primary Citation of Related Structures:
3G67, 3G6B - PubMed Abstract:
Transmembrane chemoreceptors, also known as methyl-accepting chemotaxis proteins (MCPs), translate extracellular signals into intracellular responses in the bacterial chemotaxis system. MCP ligand binding domains control the activity of the CheA kinase, situated approximately 200 A away, across the cytoplasmic membrane. The 2.17 A resolution crystal structure of a Thermotoga maritima soluble receptor (Tm14) reveals distortions in its dimeric four-helix bundle that provide insight into the conformational states available to MCPs for propagating signals. A bulge in one helix generates asymmetry between subunits that displaces the kinase-interacting tip, which resides more than 100 A away. The maximum bundle distortion maps to the adaptation region of transmembrane MCPs where reversible methylation of acidic residues tunes receptor activity. Minor alterations in coiled-coil packing geometry translate the bulge distortion to a >25 A movement of the tip relative to the bundle stalks. The Tm14 structure discloses how alterations in local helical structure, which could be induced by changes in methylation state and/or by conformational signals from membrane proximal regions, can reposition a remote domain that interacts with the CheA kinase.
Organizational Affiliation:
Department of Chemistry and Chemical Biology, Cornell University, Ithaca, New York 14850, USA.