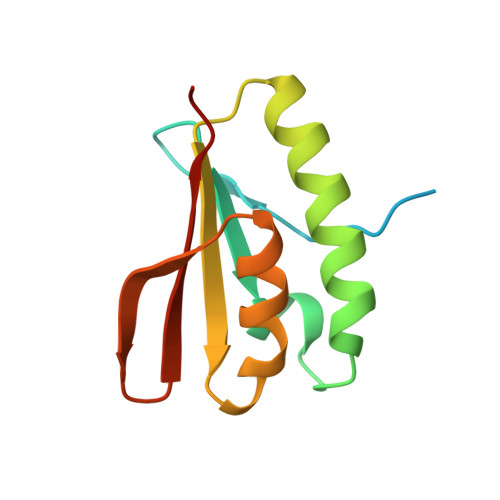
Crystal structure of the N-terminal domain of human NUDT6
Tresaugues, L., Welin, M., Arrowsmith, C.H., Berglund, H., Bountra, C., Collins, R., Dahlgren, L.G., Edwards, A.M., Flodin, S., Flores, A., Graslund, S., Hammarstrom, M., Johansson, A., Johansson, I., Karlberg, T., Kotenyova, T., Lehtio, L., Moche, M., Nilsson, M.E., Nyman, T., Persson, C., Sagemark, J., Schueler, H., Siponen, M.I., Thorsell, A.G., Van Den Berg, S., Weigelt, J., Wikstrom, M., Wisniewska, M., Nordlund, P.To be published.