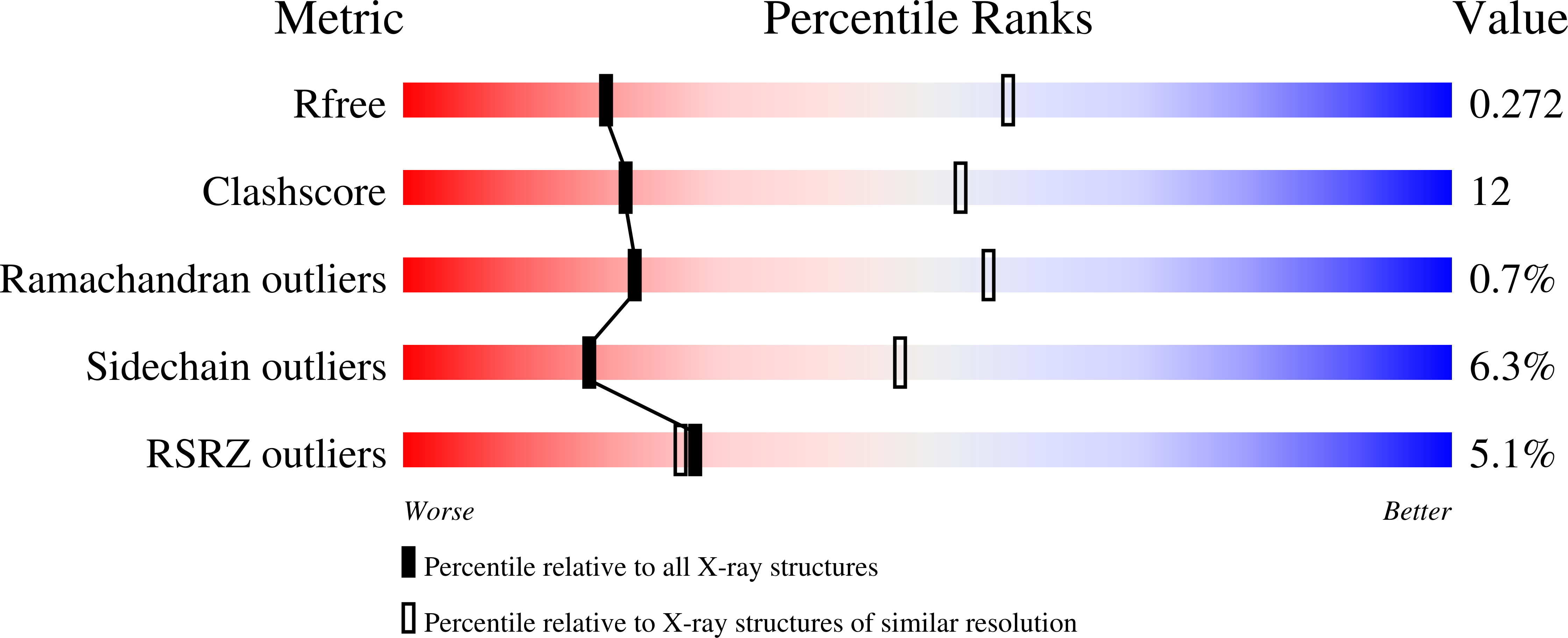
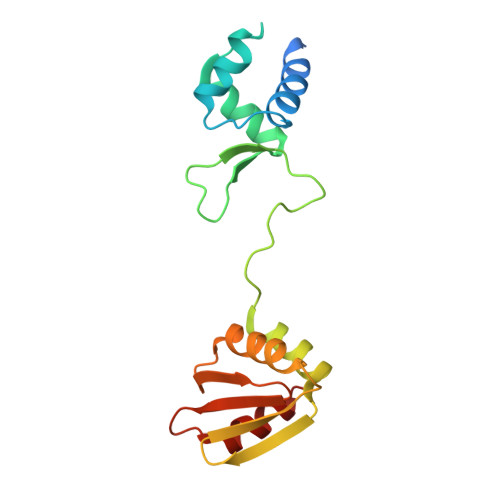
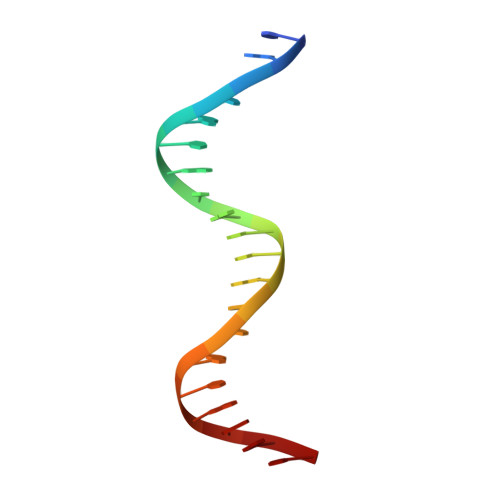
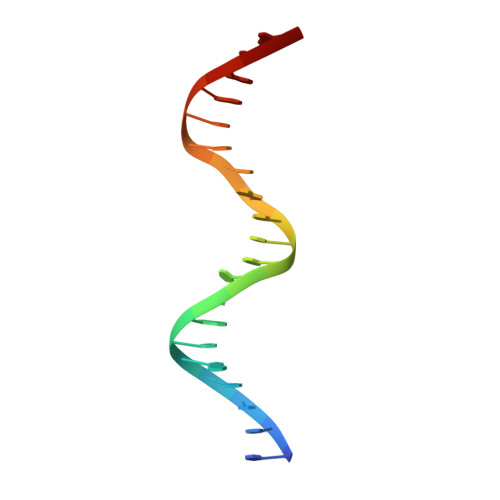
The structure of the arginine repressor from Mycobacterium tuberculosis bound with its DNA operator and Co-repressor, L-arginine.
Cherney, L.T., Cherney, M.M., Garen, C.R., James, M.N.(2009) J Mol Biol 388: 85-97
- PubMed: 19265706
- DOI: https://doi.org/10.1016/j.jmb.2009.02.053
- Primary Citation of Related Structures:
3FHZ - PubMed Abstract:
The biosynthesis of arginine is an essential function for the metabolism of Mycobacterium tuberculosis (Mtb) and for the metabolism of many other microorganisms. The arginine repressor (ArgR) proteins control the transcription of genes encoding the arginine biosynthetic enzymes; they belong to repressors having one of the most intricate oligomerization patterns. Here, we present the crystal structure of the MtbArgR hexamer bound to three copies of the 20 base-pair DNA operator and to the co-repressor, L-arginine, determined to 3.3 A resolution. This is the first ternary structure of an intact hexameric ArgR in complex with its DNA operator. The structure reported here is very different from the suggested models of the ternary ArgR-DNA complexes; it has revealed the sophisticated symmetry of the complex and the presence of two remarkably different protomer conformations, folded and extended. Both features provide flexibility to DNA binding and are important for understanding the detailed function of ArgRs. Two of the 20 base-pair DNA operators align in a unified double-helical structure, suggesting the possible presence of a double ARG box in the promoter region of the Mtb arginine operon. Two pairs of protomers bind to the unified double ARG box so that the two folded protomers bind to the central half-sites of the double ARG box, whereas the two extended protomers bind to the remote half-sites. The protomers of the third pair bound to the single DNA operator also have a folded and an extended conformation. A probable mechanism for arginine repression is suggested on the basis of this structure.
Organizational Affiliation:
Group in Protein Structure and Function, Department of Biochemistry, University of Alberta, Edmonton, Alberta, Canada T6G 2H7.