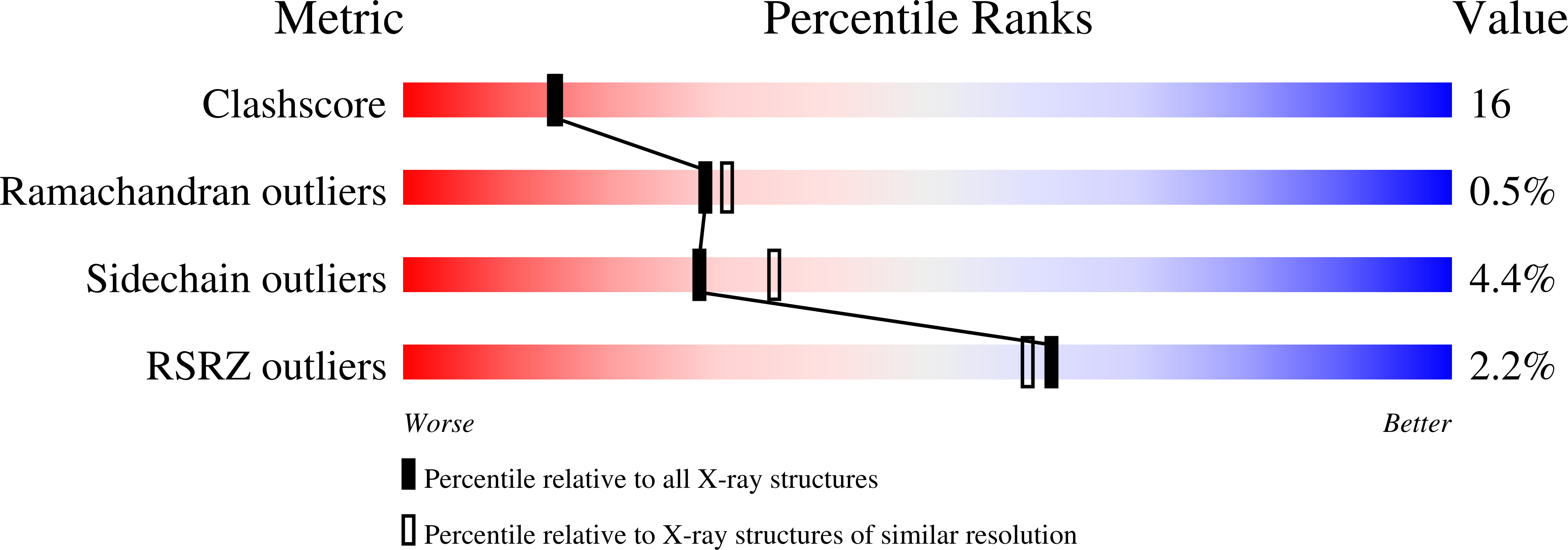Crystal structure of a ferredoxin reductase for the CYP199A2 system from Rhodopseudomonas palustris
Xu, F., Bell, S.G., Peng, Y., Johnson, E.O., Bartlam, M., Rao, Z., Wong, L.L.(2009) Proteins 77: 867-880
- PubMed: 19626710
- DOI: https://doi.org/10.1002/prot.22510
- Primary Citation of Related Structures:
3FG2 - PubMed Abstract:
Cytochrome P450-199A2 from Rhodopseudomonas palustris oxidizes para-substituted benzoic acids and may play a role in lignin and aromatic acid degradation pathways in the bacterium. CYP199A2 has an associated [2Fe-2S] ferredoxin, palustrisredoxin (Pux) but not a ferredoxin reductase. A genome search identified the palustrisredoxin reductase (PuR) gene. PuR was produced in Escherichia coli and shown to be a flavin-dependent protein that supports efficient electron transfer from NADH to Pux, thus reconstituting CYP199A2 monooxygenase activity (k(cat) = 37.9 s(-1) with 4-methoxybenzoic acid). The reduction of Pux by PuR shows K(m) = 4.2 microM and k(cat) = 262 s(-1) in 50 mM Tris, pH 7.4. K(m) is increased to 154 microM in the presence of 200 mM KCl, indicating the importance of ionic interactions in PuR/Pux binding. The crystal structure of PuR has been determined at 2.2 A resolution and found to be closely related to that of other oxygenase-coupled NADH-dependent ferredoxin reductases. Residues on the surface that had been proposed to be involved in ferredoxin reductase-ferredoxin binding are conserved in PuR. However, Lys328 in PuR lies over the FAD isoalloxazine ring and, together with His11 and Gln41, render the electrostatic potential of the surface more positive and may account for the greater involvement of electrostatic interactions in ferredoxin binding by PuR. Consistent with these observations the K328G mutation weakened Pux binding and virtually eliminated the dependence of PuR/Pux binding on salt concentration, thus confirming that the FAD si side surface in the vicinity of Lys328 is the ferredoxin binding site.
Organizational Affiliation:
Tsinghua-Nankai-IBP Joint Research Group for Structural Biology, Department of Biological Science and Biotechnology, Tsinghua University, Beijing 100084, People's Republic of China.