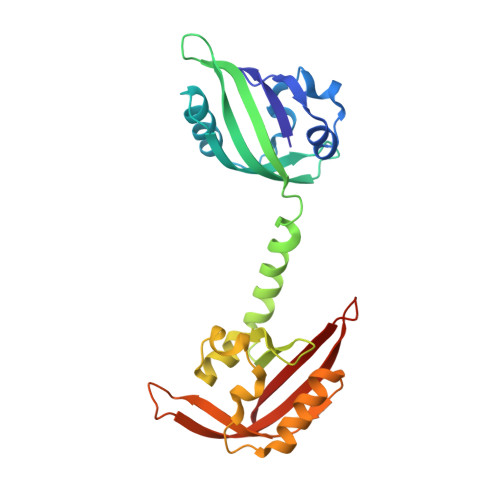
Structure of the Redox Sensor Domain of Methylococcus capsulatus (Bath) MmoS.
Ukaegbu, U.E., Rosenzweig, A.C.(2009) Biochemistry 48: 2207-2215
- PubMed: 19271777
- DOI: https://doi.org/10.1021/bi8019614
- Primary Citation of Related Structures:
3EWK - PubMed Abstract:
MmoS from Methylococcus capsulatus (Bath) is the multidomain sensor protein of a two-component signaling system proposed to play a role in the copper-mediated regulation of soluble methane monooxygenase (sMMO). MmoS binds an FAD cofactor within its N-terminal tandem Per-Arnt-Sim (PAS) domains, suggesting that it functions as a redox sensor. The crystal structure of the MmoS tandem PAS domains, designated PAS-A and PAS-B, has been determined to 2.34 A resolution. Both domains adopt the typical PAS domain alpha/beta topology and are structurally similar. The two domains are linked by a long alpha helix and do not interact with one another. The FAD cofactor is housed solely within PAS-A and is stabilized by an extended hydrogen bonding network. The overall fold of PAS-A is similar to those of other flavin-containing PAS domains, but homodimeric interactions in other structures are not observed in the MmoS sensor, which crystallized as a monomer. The structure both provides new insight into the architecture of tandem PAS domains and suggests specific residues that may play a role in MmoS FAD redox chemistry and subsequent signal transduction.
Organizational Affiliation:
Department of Biochemistry, Northwestern University, Evanston, Illinois 60208, USA.