Radiation damage in protein structural characterization by Synchrotron Radiation: State of the art and Nanotechnology-based perspective
Pechkova, E., Tripathi, S.K., Nicolini, C.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Thermolysin | 316 | Bacillus thermoproteolyticus | Mutation(s): 0 EC: 3.4.24.27 | 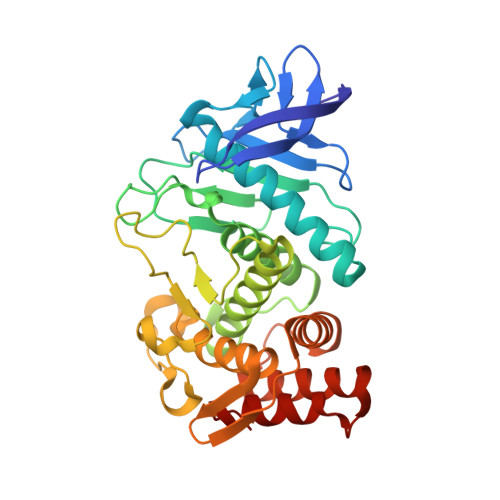 | |
UniProt | |||||
Find proteins for P00800 (Bacillus thermoproteolyticus) Explore P00800 Go to UniProtKB: P00800 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P00800 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| LYS Query on LYS | H [auth A] | LYSINE C6 H15 N2 O2 KDXKERNSBIXSRK-YFKPBYRVSA-O |  | ||
| VAL Query on VAL | G [auth A] | VALINE C5 H11 N O2 KZSNJWFQEVHDMF-BYPYZUCNSA-N |  | ||
| ZN Query on ZN | F [auth A] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| CA Query on CA | B [auth A], C [auth A], D [auth A], E [auth A] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 92.724 | α = 90 |
| b = 92.724 | β = 90 |
| c = 128.6 | γ = 120 |
| Software Name | Purpose |
|---|---|
| MOSFLM | data reduction |
| SCALA | data scaling |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |