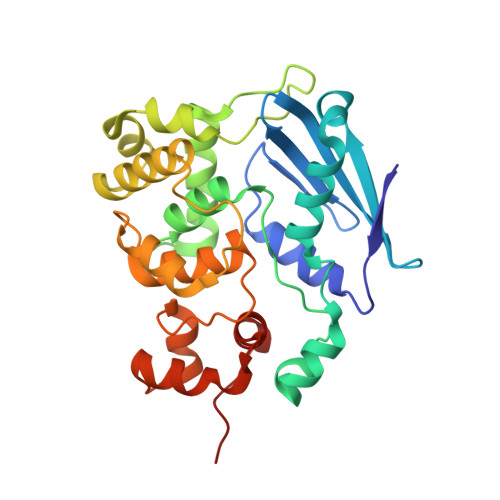
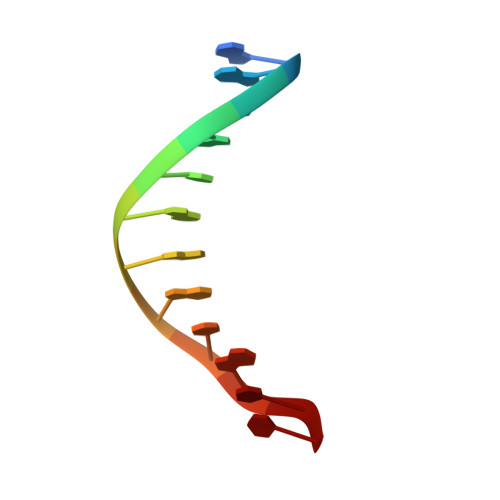
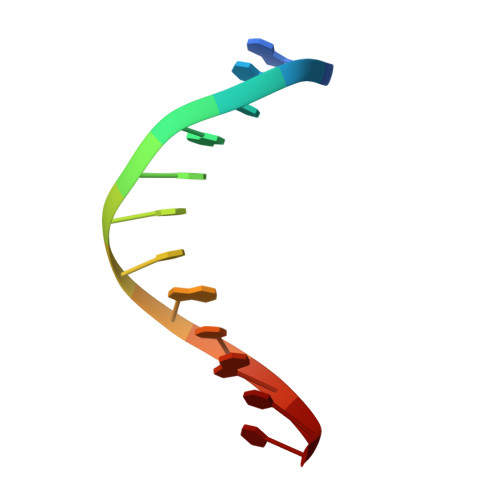
Structure of the E. coli DNA Glycosylase AlkA Bound to the Ends of Duplex DNA: A System for the Structure Determination of Lesion-Containing DNA.
Bowman, B.R., Lee, S., Wang, S., Verdine, G.L.(2008) Structure 16: 1166-1174
- PubMed: 18682218
- DOI: https://doi.org/10.1016/j.str.2008.04.012
- Primary Citation of Related Structures:
3CVS, 3CVT, 3CW7, 3CWA, 3CWS, 3CWT, 3CWU - PubMed Abstract:
The constant attack on DNA by endogenous and exogenous agents gives rise to nucleobase modifications that cause mutations, which can lead to cancer. Visualizing the effects of these lesions on the structure of duplex DNA is key to understanding their biologic consequences. The most definitive method of obtaining such structures, X-ray crystallography, is troublesome to employ owing to the difficulty of obtaining diffraction-quality crystals of DNA. Here, we present a crystallization system that uses a protein, the DNA glycosylase AlkA, as a scaffold to mediate the crystallization of lesion-containing duplex DNA. We demonstrate the use of this system to facilitate the rapid structure determination of DNA containing the lesion 8-oxoguanine in several different sequence contexts, and also deoxyinosine and 1,N(6)-ethenoadenine, each stabilized as the corresponding 2'-flouro analog. The structures of 8-oxoguanine provide a correct atomic-level view of this important endogenous lesion in DNA.
Organizational Affiliation:
Department of Chemistry and Chemical Biology, Harvard University, Cambridge, MA 02138, USA.