X-ray Structures of the Three Lactococcus lactis Dihydroxyacetone Kinase Subunits and of a Transient Intersubunit Complex.
Zurbriggen, A., Jeckelmann, J.M., Christen, S., Bieniossek, C., Baumann, U., Erni, B.(2008) J Biol Chem 283: 35789-35796
- PubMed: 18957416
- DOI: https://doi.org/10.1074/jbc.M804893200
- Primary Citation of Related Structures:
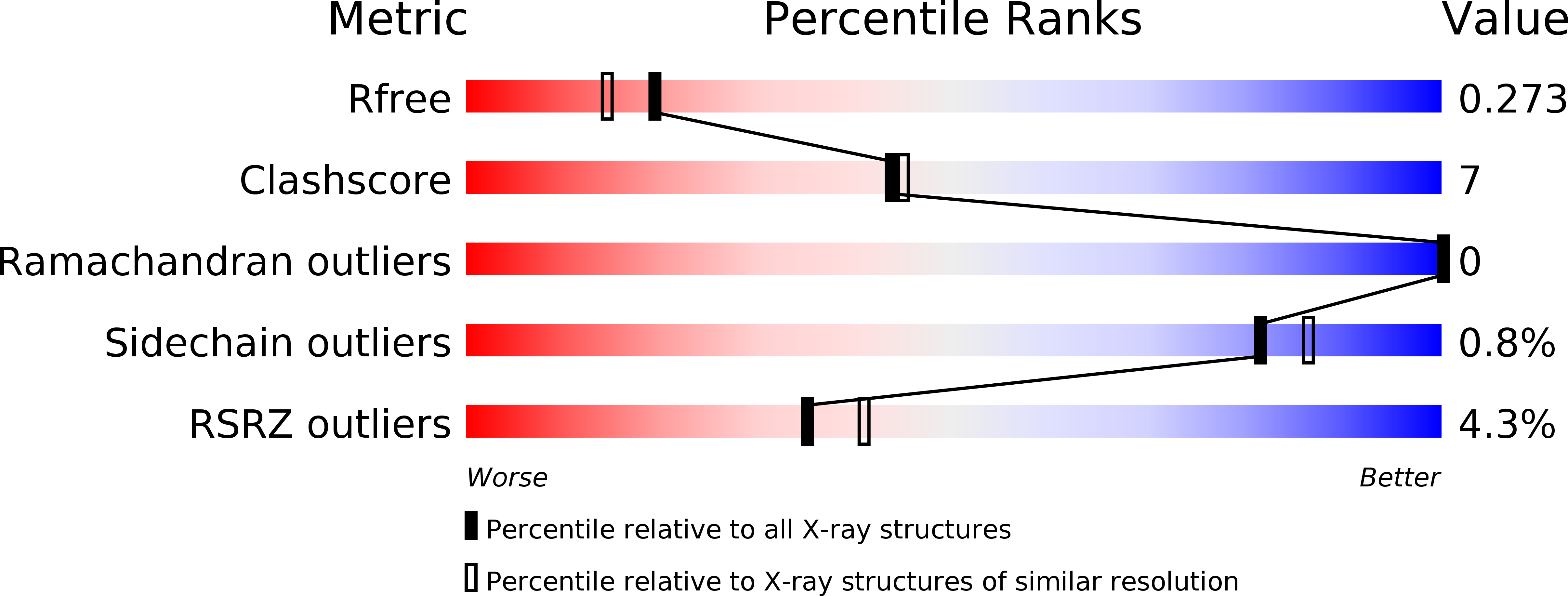
3CR3, 3CT4, 3CT6 - PubMed Abstract:
Bacterial dihydroxyacetone (Dha) kinases do not exchange the ADP for ATP but utilize a subunit of the phosphoenolpyruvate carbohydrate phosphotransferase system for in situ rephosphorylation of a permanently bound ADP-cofactor. Here we report the 2.1-angstroms crystal structure of the transient complex between the phosphotransferase subunit DhaM of the phosphotransferase system and the nucleotide binding subunit DhaL of the Dha kinase of Lactococcus lactis, the 1.1-angstroms structure of the free DhaM dimer, and the 2.5-angstroms structure of the Dha-binding DhaK subunit. Conserved salt bridges and an edge-to-plane stacking contact between two tyrosines serve to orient DhaL relative to the DhaM dimer. The distance between the imidazole Nepsilon2 of the DhaM His-10 and the beta-phosphate oxygen of ADP, between which the gamma-phosphate is transferred, is 4.9 angstroms. An invariant arginine, which is essential for activity, is appropriately positioned to stabilize the gamma-phosphate in the transition state. The (betaalpha)4alpha fold of DhaM occurs a second time as a subfold in the DhaK subunit. By docking DhaL-ADP to this subfold, the nucleotide bound to DhaL and the C1-hydroxyl of Dha bound to DhaK are positioned for in-line transfer of phosphate.
Organizational Affiliation:
Departement für Chemie und Biochemie, Universität Bern, Freiestrasse 3, Bern CH-3012, Switzerland.